GO.db
相似的基因在不同物种中,其功能往往保守的。显然,需要一个统一的术语用于描述这些跨物种的同源基因及其基因产物的功能,否则,不同的实验室对相同的基因的功能的描述不同,将极大限制学术的交流。而 Gene Ontology (GO) 项目正是为了能够使对各种数据库中基因获基因产物功能描述相一致的努力结果。一般情况下,在各种组学中,做完差异表达之后,会看这些差异表达基因参与的功能,一般我们会做一个GO注释和富集。这里我们先来了解一下GO.db这个包。因为该包是许多其他包的基础依赖包。对于了解GO有很大的帮助。
1)安装载入
-------------------------------------------
if("GO.db" %in% rownames(installed.packages()) == FALSE) {source("http://bioconductor.org/biocLite.R");biocLite("GO.db")}
suppressMessages(library(GO.db))
2)查看该包所有的对象
--------------------------------------------
ls("package:GO.db")
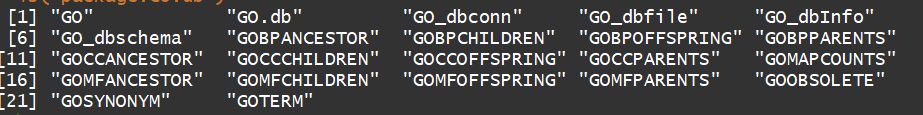
columns(GO.db)
"DEFINITION" "GOID" "ONTOLOGY" "TERM"
功能:可以用来进行GO的注释
GOBPANCESTOR:Annotation of GO Identifiers to their Biological Process Ancestors
GOBPCHILDREN:Annotation of GO Identifiers to their Biological Process Children
GOBPOFFSPRING: Annotation of GO Identifiers to their Biological Process Offspring
GOBPPARENTS: Annotation of GO Identifiers to their Biological Process Parents
GOCCANCESTOR: Annotation of GO Identifiers to their Cellular Component Ancestors
GOCCCHILDREN: Annotation of GO Identifiers to their Cellular Component Children
GOCCOFFSPRING: Annotation of GO Identifiers to their Cellular Component Offspring
GOCCPARENTS: Annotation of GO Identifiers to their Cellular Component Parents
GOMAPCOUNTS Number of mapped keys for the maps in package GO.db
GOMFANCESTOR: Annotation of GO identifiers to their Molecular Function Ancestors
GOMFCHILDREN: Annotation of GO Identifiers to their Molecular Function Children
GOMFOFFSPRING: Annotation of GO Identifiers to their Molecular Function Offspring
GOMFPARENTS: Annotation of GO Identifiers to their Molecular Function Parents
GOOBSOLETE: Annotation of GO identifiers by terms defined by Gene Ontology Consortium and their status are obsolete
GOSYNONYM:Map from GO synonyms to GO terms
GOTERM: Annotation of GO Identifiers to GO Terms
GO_dbconn:Collect information about the package: Annotation DB
原理:所依赖的数据库ftp://ftp.geneontology.org/pub/go/godatabase/archive/latest-lite/
3)各个对象的简单使用
-----------------------------------------------------------
3.1)GOBPANCESTOR (Annotation of GO Identifiers to all of their Biological Process Ancestors,即返回的是该GO号所有的长辈关系)
xx <- as.list(GOBPANCESTOR) # Convert the object to a list
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any ancestor
if(length(xx) > 0){
goids <- xx[1] # Get the ancestor GO IDs for the first two elents of xx
}
goids
$`GO:0000001`
[1] "GO:0006996" "GO:0007005" "GO:0008150" "GO:0009987" "GO:0016043" "GO:0048308"
[7] "GO:0048311" "GO:0051179" "GO:0051640" "GO:0051641" "GO:0051646" "GO:0071840"
[13] "all"
3.2)GOBPCHILDREN (Annotation of GO Identifiers to their Biological Process direct Children,即返回的是 嫡系儿子)
xx <- as.list(GOBPCHILDREN)
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any children
if(length(xx) > 0){
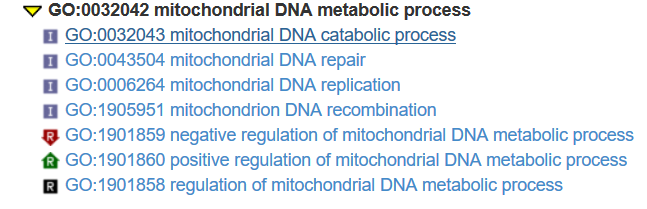
goids <- xx[[1]] # Get the parent GO IDs for the first elents of xx part_of "GO:0032042" ; is_a "GO:0033955"
GOID(GOTERM[[goids[1]]]) #"GO:0032042"
Term(GOTERM[[goids[1]]]) #"mitochondrial DNA metabolic process"
Synonym(GOTERM[[goids[1]]]) #"mitochondrial DNA metabolism" "mtDNA metabolic process" "mtDNA metabolism"
Secondary(GOTERM[[goids[1]]]) #character(0)
Definition(GOTERM[[goids[1]]]) # "The chemical reactions and pathways involving mitochondrial DNA."
Ontology(GOTERM[[goids[1]]]) #"BP"
}
3.3)GOBPOFFSPRING(Annotation of GO Identifiers to their Biological Process Offspring,子孙后裔,即相当于所有有血缘关系)
xx <- as.list(GOBPOFFSPRING)
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any offspring
if(length(xx) > 0){
goids <- xx[1] # Get the offspring GO IDs for the first of xx
}
goids
$`GO:0000002`
[1] "GO:0006264" "GO:0032042" "GO:0032043" "GO:0033955" "GO:0043504" "GO:0090296"
[7] "GO:0090297" "GO:0090298" "GO:1901858" "GO:1901859" "GO:1901860" "GO:1905951"

3.4) GOBPPARENTS(Annotation of GO Identifiers to their Biological Process direct Parents,即返回的是直接父亲)
xx <- as.list(GOBPPARENTS)
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any parent
if(length(xx) > 0){
goids <- xx[[1]] # Get the children GO IDs for the first elents of xx #is_a "GO:0048308" ,is_a "GO:0048311"
GOID(GOTERM[[goids[1]]]) # Find out the GO terms for the first parent goid #"GO:0048308"
Term(GOTERM[[goids[1]]]) #"organelle inheritance"
Synonym(GOTERM[[goids[1]]]) #character(0)
Secondary(GOTERM[[goids[1]]]) #character(0)
Definition(GOTERM[[goids[1]]]) #"The partitioning of organelles between daughter cells at cell division."
Ontology(GOTERM[[goids[1]]]) #"BP"
}
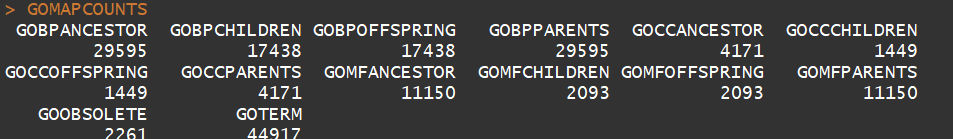
3.5)GOMAPCOUNTS (Number of mapped keys for the maps in package GO.db)
GOMAPCOUNTS
3.6)GOOBSOLETE(Annotation of GO identifiers by terms defined by Gene Ontology Consortium and their status are obsolete)
xx <- as.list(GOTERM)
if(length(xx) > 0){
GOID(xx[[1]]) # Get the TERMS for the first elent of xx "GO:0000001"
Ontology(xx[[1]]) "BP"
}
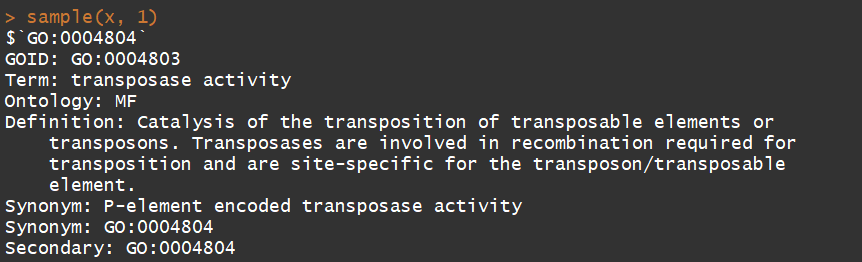
3.7)GOSYNONYM(Map from GO synonyms to GO terms)
x <- GOSYNONYM
sample(x, 1)
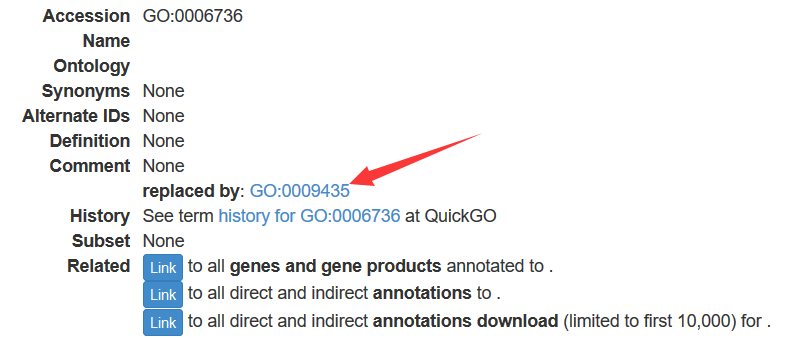
GOTERM[["GO:0009435"]] # GO ID "GO:0009435" has a lot of synonyms
GOID(GOSYNONYM[["GO:0006736"]]) # GO ID "GO:0006736" is a synonym of GO ID "GO:0009435"
3.8)GOTERM Annotation of GO Identifiers to GO Terms
xx <- as.list(GOTERM)
if(length(xx) > 0){
GOID(xx[[1]]) ##"GO:0000001"
Term(xx[[1]]) ##"mitochondrion inheritance"
Synonym(xx[[1]]) #"mitochondrial inheritance"
Secondary(xx[[1]]) ##character(0)
Definition(xx[[1]]) #"The distribution of mitochondria, including the mitochondrial genome, into daughter cells after mitosis or meiosis, mediated by interactions between mitochondria and the cytoskeleton.
Ontology(xx[[1]]) #"BP"
}
3.9)GO_dbconn(Collect information about the package annotation DB)
GO_dbconn()
GO_dbfile()
GO_dbInfo()
这里以BP为例,CC,MF和其它道理相同
GO.db的更多相关文章
- Linux平台 Oracle 10gR2(10.2.0.5)RAC安装 Part3:db安装和升级
Linux平台 Oracle 10gR2(10.2.0.5)RAC安装 Part3:db安装和升级 环境:OEL 5.7 + Oracle 10.2.0.5 RAC 5.安装Database软件 5. ...
- VS15 preview 5打开文件夹自动生成slnx.VC.db SQLite库疑惑?求解答
用VS15 preview 5打开文件夹(详情查看博客http://www.cnblogs.com/zsy/p/5962242.html中配置),文件夹下多一个slnx.VC.db文件,如下图: 本文 ...
- ODBC、OLE DB、 ADO的区别
转自:http://blog.csdn.net/yinjingjing198808/article/details/7665577 一.ODBC ODBC的由来 1992年Microsoft和Syba ...
- Linux平台 Oracle 11gR2 RAC安装Part3:DB安装
四.DB(Database)安装 4.1 解压DB的安装包 4.2 DB软件安装 4.3 ASMCA创建磁盘组 4.4 DBCA建库 4.5 验证crsctl的状态 Linux平台 Oracle 11 ...
- SSRS ----环境配置,没有 ReportServer DB 怎么办?
今天项目进入报表开发阶段,按照习惯,打开报表管理器,发现提示下面的错误: 错误:报表服务器无法打开与报表服务器数据库的连接.所有请求和处理都要求与数据库建立连接. 这是怎么回事儿呢,经过排查,发现数据 ...
- mongo DB for C#
(1)Download the MongoDB C#驱动. http://www.nuget.org/packages/mongocsharpdriver/. (2) Add Reference to ...
- jBPM4.4 no jBPM DB schema: no JBPM4_EXECUTION table. Run the create.jbpm.schema target first in the install tool.
jBPM4.4 no jBPM DB schema: no JBPM4_EXECUTION table. Run the create.jbpm.schema target first in the ...
- oracle db link的查看创建与删除
1.查看dblink select owner,object_name from dba_objects where object_type='DATABASE LINK'; 或者 select * ...
- Gc.Db之循序渐进
距离上次写Gc.Db框架已经有一段时间了,最近默默对框架代码已经做了不少优化和功能,且已经提交至nuget,大家如果想使用此框架,可以通过nuget搜索:Gc.Db进行下载和安装包. 本篇文章主要是介 ...
- oracle Entity db.Database.SqlQuery ORA-01843: 无效的月份
原因是oracle的日期格式化格式和本地语言环境的日期格式不一致导致的. 一般情景为oralce格式为英文格式 本地服务器或者开发机的环境为中文环境. 使用Dbcontext 实例一般不会有问题. 但 ...
随机推荐
- dedecms问答模块时间显示1970-01-01的解决办法
在使用dedecms的问答模块的时候,很多人希望能在模板上加上问题的时间,一般我们采用这样的dede标签: {dede:ask row=24 titlelen='50' sort='ok'} &l ...
- <<APUE>> 线程
一个进程在同一时刻只能做一件事情,线程可以把程序设计成在同一时刻能够做多件事情,每个线程处理各自独立的任务.线程包括了表示进程内执行环境必需的信息,包括进程中标识线程的线程ID.一组寄存器值.栈.调度 ...
- ssl证书(https) iis 配置安装
因客户给的 cer的文件 导入提示 失败,所以用 了 客户给的 crt的格式的证书. 安装证书操作如下:iis>>服务器证书>>右侧菜单-完成证书申请>>选择 本文 ...
- bzoj4928: 第二题
Description 对于一棵有根树,定义一个点u的k-子树为u的子树中距离u不超过k的部分. 注意,假如u的子树中不存在距离u为k的点,则u的k-子树是不存在的. 定义两棵子树是相同的,当且仅当不 ...
- PAT 乙级 1037 在霍格沃茨找零钱(20)C++版
1037. 在霍格沃茨找零钱(20) 时间限制 400 ms 内存限制 65536 kB 代码长度限制 8000 B 判题程序 Standard 作者 CHEN, Yue 如果你是哈利·波特迷,你会知 ...
- Open Live writer 远程博客管理客户端
1. 官网地址:http://openlivewriter.org/ 点击download下载:https://openlivewriter.azureedge.net/stable/Release ...
- freePBX汉化方法记录——备忘
FreePBX汉化[root@bgcc69:/var/www/html/admin/i18n/zh_CN/LC_MESSAGES]$pwd/var/www/html/admin/i18n/zh_CN/ ...
- 补充2:Golang 一些特性
Go语言的这些地方都做的还不错: 拥有自动垃圾回收: 不用手动释放内存 一个包系统: Go 语言的源码复用建立在包(package)基础之上.包通过 package, import, GOPATH 操 ...
- ossim 4.1 安装
在这说明一下,本人也是第一次安装这东东很多我自己也不懂,只能把自己的安装步骤通过贴图呈现出来了!!!! 我用root用户登录的,顺便查看ip 在浏览器打开网址https://192.168.86.15 ...
- Spark standalone运行模式
Spark Standalone 部署配置 Standalone架构 手工启动一个Spark集群 https://spark.apache.org/docs/latest/spark-standalo ...