org.Hs.eg.db包简介(转换NCBI、ensemble等数据库中基因ID,symbol等之间的转换)
1)安装载入
-------------------------------------------
if("org.Hs.eg.db" %in% rownames(installed.packages()) == FALSE) {source("http://bioconductor.org/biocLite.R");biocLite("org.Hs.eg.db")}
suppressMessages(library(org.Hs.eg.db))
2)查看该包所有的对象
--------------------------------------------
ls("package:org.Hs.eg.db")
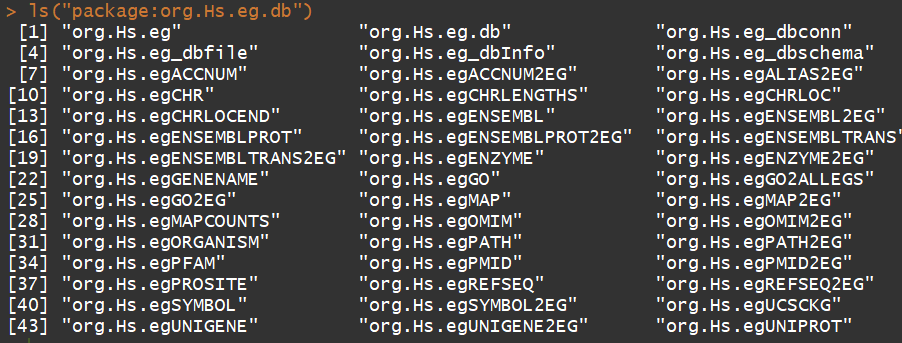
功能:可以用来进行基因ID的转换
org.Hs.egACCNUM:Map Entrez Gene identifiers to GenBank Accession Numbers(Entrez Gene identifiers 和genbank)
org.Hs.egALIAS2EG:Map between Common Gene Symbol Identifiers and Entrez Gene
org.Hs.eg.db:Bioconductor annotation data package
org.Hs.egCHR:Map Entrez Gene IDs to Chromosomes
org.Hs.egCHRLENGTHS:A named vector for the length of each of the chromosomes
org.Hs.egCHRLOC:Entrez Gene IDs to Chromosomal Location
org.Hs.egENSEMBL:Map Ensembl gene accession numbers with Entrez Gene identifiers
org.Hs.egENSEMBLPROT:Map Ensembl protein acession numbers with Entrez Gene identifiers
org.Hs.egENSEMBLTRANS:Map Ensembl transcript acession numbers with Entrez Gene identifiers
org.Hs.egENZYME:Map between Entrez Gene IDs and Enzyme Commission (EC) Numbers
org.Hs.egGENENAME:Map between Entrez Gene IDs and Genes
org.Hs.egGO:Maps between Entrez Gene IDs and Gene Ontology (GO) IDs
org.Hs.egMAP:Map between Entrez Gene Identifiers and cytogenetic:Maps/bands
org.Hs.egMAPCOUNTS Number of:Mapped keys for the:Maps in package org.Hs.eg.db
org.Hs.egOMIM:Map between Entrez Gene Identifiers and Mendelian Inheritance in Man (MIM) identifiers
org.Hs.egORGANISM:The Organism for org.Hs.eg
org.Hs.egPATH:Mappings between Entrez Gene identifiers and KEGG pathway identifiers
org.Hs.egPFAM:Maps between Manufacturer Identifiers and PFAM Identifiers
org.Hs.egPMID:Map between Entrez Gene Identifiers and PubMed Identifiers
org.Hs.egPROSITE:Maps between Manufacturer Identifiers and PROSITE Identifiers
org.Hs.egREFSEQ:Map between Entrez Gene Identifiers and RefSeq Identifiers
org.Hs.egSYMBOL:Map between Entrez Gene Identifiers and Gene Symbols
org.Hs.egUNIGENE:Map between Entrez Gene Identifiers and UniGene cluster identifiers
org.Hs.egUNIPROT:Map Uniprot accession numbers with Entrez Gene identifiers
org.Hs.eg_dbconn:Collect information about the package annotation DB
示例:
(用mget函数):
myEIDs <- c("1", "10", "100", "1000", "37690")
mySymbols <- mget(myEIDs, org.Hs.egSYMBOL, ifnotfound=NA) ####myEID是自己的ID,org.Hs.egSYMBOL是其中的一个对象
mySymbols <- unlist(mySymbols)
(用select函数):
myEIDs <- c("ENSG00000130720", "ENSG00000103257", "ENSG00000156414")
cols <- c("SYMBOL", "GENENAME")
select(org.Hs.eg.db, keys=myEIDs, columns=cols, keytype="ENSEMBL")#生成数据框,
原理:例如将 Entrez Gene identifiers( https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene) 与 GenBank accession numbers进行简单的mapping。该map依据的数据库是Entrez Gene ftp://ftp.ncbi.nlm.nih.gov/gene/DATA
以DATA其中的一个gene2ensembl文件为例来感受其实如何实现的:
wget ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2ensembl.gz
解压后查看:
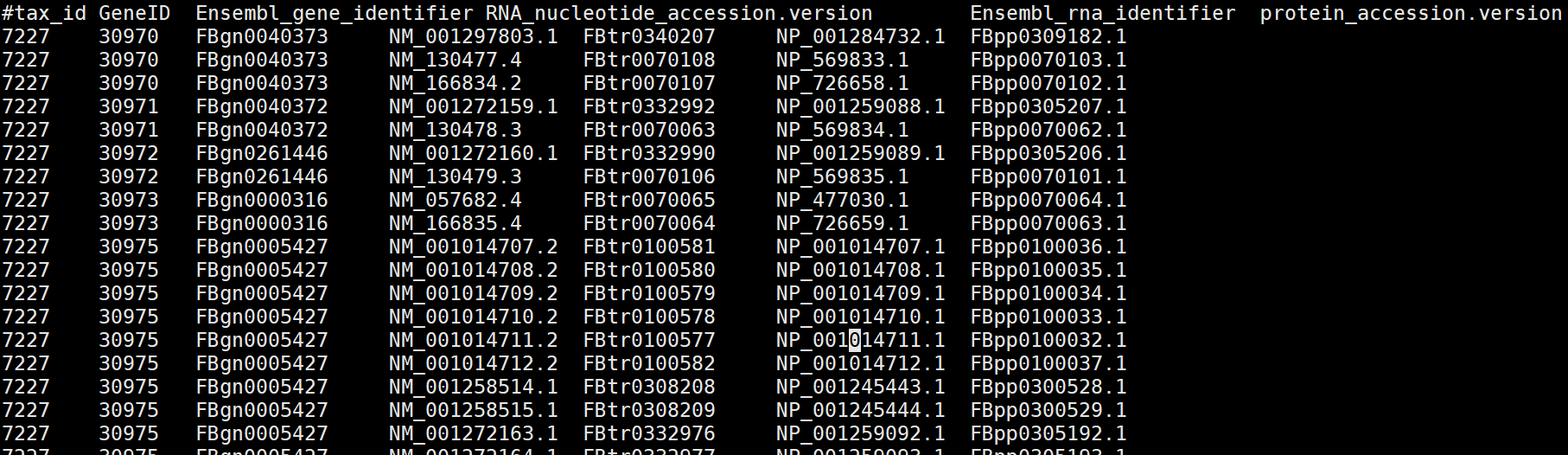
其中第一列是物种id,第二列是GeneID, 第三列是Ensemble_geneID,第四列是RNA_id,第五列是Ensemble_RNAid,第六列是protein_id。因此这些R包的功能极有可能就是利用NCBI或ensem等数据库中的这些文件信息,通过一系列的脚本实现了基因ID之间进行转换,因此如果对NCBI、Ensemble等网络架构熟悉的话,自己又会写脚本,就可以自己处理,而不用这些R包进行。当然别人写好了,为什么自己造轮子呢?自己造轮子是为了深刻的理解
3)各个对象的简单使用
-----------------------------------------------------------
3.1)org.Hs.egACCNUM(将Entrez Gene identifiers 与 GenBank Accession Numbers进行map
x <- org.Hs.egACCNUM ### Bimap interface
mapped_genes <- mappedkeys(x) ## Get the entrez gene identifiers that are mapped to an ACCNUM
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the ACCNUM for the first five genes
xx[[1]] # Get the first one
}
#For the reverse map ACCNUM2EG:
xx <- as.list(org.Hs.egACCNUM2EG) # Convert to a list
if(length(xx) > 0){
xx[1:5] # Gets the entrez gene identifiers for the first five Entrez Gene IDs
xx[[1]] # Get the first one
}
3.2)org.Hs.egALIAS2EG(将 Common Gene Symbol Identifiers 和 Entrez Gene进行转换)
x <- org.Hs.egACCNUM ## Bimap interface:org.Hs.egALIAS2EG
xx <- as.list(org.Hs.egALIAS2EG) # Convert the object to a list
xx <- xx[!is.na(xx)] # Remove pathway identifiers that do not map to any entrez gene id
if(length(xx) > 0){
xx[1:2] # The entrez gene identifiers for the first two elements of XX
xx[[1]] # Get the first one
}
3.3) org.Hs.egCHR (将Entrez Gene IDs 和Chromosomes进行map)
x <- org.Hs.egCHR ## Bimap interface
mapped_genes <- mappedkeys(x) #Get entrez gene that are mapped to a chromosome
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the CHR for the first five genes
xx[[1]] # Get the first one
}
3.4)org.Hs.egCHRLENGTHS (每个染色体的长度)
tt <- org.Hs.egCHRLENGTHS ## Bimap interface:
tt["1"] # Length of chromosome 1
for (i in c(1:22,'X','Y')){print(tt[i])} #####打印每一个染色体的长度
3.5) org.Hs.egCHRLOC (Entrez Gene IDs在Chromosomal 上的定位)
x <- org.Hs.egCHRLOC ### Bimap interface
mapped_genes <- mappedkeys(x) #Get the entrez gene identifiers that are mapped to chromosome locations
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the CHRLOC for the first five genes
xx[[1]] # Get the first one
}
3.6)org.Hs.egENSEMBL (将Ensembl gene accession numbers 与 Entrez Gene identifiers进行map)
x <- org.Hs.egENSEMBL ## Bimap interface
mapped_genes <- mappedkeys(x)# Get the entrez gene IDs that are mapped to an Ensembl ID
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the Ensembl gene IDs for the first five genes
xx[[1]] # Get the first one
}
#For the reverse map ENSEMBL2EG:
xx <- as.list(org.Hs.egENSEMBL2EG) # Convert to a list
if(length(xx) > 0){
xx[1:5] # Gets the entrez gene IDs for the first five Ensembl IDs
xx[[1]] # Get the first one
}
3.7) org.Hs.egENSEMBLPROT (将Ensembl protein acession numbers 和 Entrez Gene identifiers进行map)
x <- org.Hs.egENSEMBLPROT ## Bimap interface
mapped_genes <- mappedkeys(x) #Get the entrez gene IDs that are mapped to an Ensembl ID
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the Ensembl gene IDs for the first five proteins
xx[[1]] # Get the first one
}
#For the reverse map ENSEMBLPROT2EG:
xx <- as.list(org.Hs.egENSEMBLPROT2EG) # Convert to a list
if(length(xx) > 0){
xx[1:5] # Gets the entrez gene IDs for the first five Ensembl IDs
xx[[1]] # Get the first one
}
3.8) org.Hs.egENSEMBLTRANS (将 Ensembl transcript acession numbers 与 Entrez Gene identifiers进行mapping)
x <- org.Hs.egENSEMBLTRANS ## Bimap interface:
mapped_genes <- mappedkeys(x) #entrez gene IDs that are mapped to an Ensembl ID
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the Ensembl gene IDs for the first five proteins
xx[[1]] # Get the first one
}
#For the reverse map ENSEMBLTRANS2EG:
xx <- as.list(org.Hs.egENSEMBLTRANS2EG) # Convert to a list
if(length(xx) > 0){
xx[1:5] # Gets the entrez gene IDs for the first five Ensembl IDs
xx[[1]] # Get the first one
}
3.9)org.Hs.egGENENAME(将 Entrez Gene IDs 与 Genes进行mapping)
x <- org.Hs.egGENENAME ## Bimap interface
mapped_genes <- mappedkeys(x) #gene names that are mapped to an entrez gene identifier
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
xx[1:5] # Get the GENE NAME for the first five genes
xx[[1]] # Get the first one
}
3.10)org.Hs.egGO (Entrez Gene IDs与 Gene Ontology (GO) IDs进行mapping)
x <- org.Hs.egGO ## Bimap interface:
mapped_genes <- mappedkeys(x) # entrez gene identifiers that are mapped to a GO ID
xx <- as.list(x[mapped_genes]) # Convert to a list
if(length(xx) > 0) {
got <- xx[[1]] # Try the first one
got[[1]][["GOID"]]
got[[1]][["Ontology"]]
got[[1]][["Evidence"]]
}
# For the reverse map:
xx <- as.list(org.Hs.egGO2EG) # Convert to a list
if(length(xx) > 0){
goids <- xx[2:3] # Gets the entrez gene ids for the top 2nd and 3nd GO identifiers
goids[[1]] # Gets the entrez gene ids for the first element of goids
names(goids[[1]]) # Evidence code for the mappings
}
# For org.Hs.egGO2ALLEGS
xx <- as.list(org.Hs.egGO2ALLEGS)
if(length(xx) > 0){
goids <- xx[2:3] # Entrez Gene identifiers for the top 2nd and 3nd GO identifiers
goids[[1]] # Gets all the Entrez Gene identifiers for the first element of goids
names(goids[[1]]) # Evidence code for the mappings
}
3.11)org.Hs.egPATH (将Entrez Gene identifiers 与KEGG pathway identifiers进行mapping)
x <- org.Hs.egPATH ## Bimap interface:
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
# For the reverse map:
xx <- as.list(org.Hs.egPATH2EG)
xx <- xx[!is.na(xx)] # Remove pathway identifiers that do not map to any entrez gene id
if(length(xx) > 0){
xx[1:2]
xx[[1]]
}
3.12)org.Hs.egREFSEQ(将Entrez Gene Identifiers 与 RefSeq Identifiers进行mapping)
x <- org.Hs.egREFSEQ
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
# For the reverse map:
x <- org.Hs.egREFSEQ2EG
mapped_seqs <- mappedkeys(x)
xx <- as.list(x[mapped_seqs])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
3.13)org.Hs.egSYMBOL(将 Entrez Gene Identifiers 与Gene Symbols进行mapping)
x <- org.Hs.egSYMBOL
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
x <- org.Hs.egSYMBOL2EG
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
3.14)org.Hs.egUNIGENE (Entrez Gene Identifiers 与 UniGene cluster identifiers进行mapping)
x <- org.Hs.egUNIGENE
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
# For the reverse map:
x <- org.Hs.egUNIGENE2EG
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
3.15)org.Hs.egUNIPROT (Uniprot accession numbers与 Entrez Gene identifiers进行mapping)
x <- org.Hs.egUNIPROT
mapped_genes <- mappedkeys(x)
xx <- as.list(x[mapped_genes])
if(length(xx) > 0) {
xx[1:5]
xx[[1]]
}
希望大家通过上述教程的解析,能够理解,基因ID,名称等之间是如何转换,并通过这些对NCBI、ensemble、pfam等数据库有相应的一定认识。
org.Hs.eg.db包简介(转换NCBI、ensemble等数据库中基因ID,symbol等之间的转换)的更多相关文章
- Oracle数据库中日期/数字和字符之间的转换和计算
--查出当前系统时间 select SYSDATE from table; --格式转换 -- TO_CHAR 把日期或数字转换为字符串 -- TO_CHAR(number, '格式') -- TO_ ...
- MFC中char*,string和CString之间的转换
MFC中char*,string和CString之间的转换 一. 将CString类转换成char*(LPSTR)类型 方法一,使用强制转换.例如: CString theString( &q ...
- C# 中List<T>与DataSet之间的转换
p{ text-align:center; } blockquote > p > span{ text-align:center; font-size: 18px; color: #ff0 ...
- 【转】Android中dip(dp)与px之间单位转换
Android中dip(dp)与px之间单位转换 dp这个单位可能对web开发的人比较陌生,因为一般都是使用px(像素)但是,现在在开始android应用和游戏后,基本上都转换成用dp作用为单位了,因 ...
- shell 脚本文件十六进制转化为ascii码代码, Shell中ASCII值和字符之间的转换
Shell中ASCII值和字符之间的转换 1.ASCII值转换为字符 方法一: i=97 echo $i | awk '{printf("%c", $1)}' ...
- Oracle中的数据类型和数据类型之间的转换
Oracle中的数据类型 /* ORACLE 中的数据类型: char 长度固定 范围:1-2000 VARCHAR2 长度可变 范围:1-4000 LONG 长度可变 最大的范围2gb 长字符类型 ...
- Java中String与Date格式之间的转换
转自:https://blog.csdn.net/angus_17/article/details/7656631 经常遇到string和date之间的转换,把相关的内容总结在这里吧: 1.strin ...
- Java中二进制数与整型之间的转换
import java.io.*; public class Test{ /** * 二进制与整型之间的转换 * @param args * @throws IOException */ public ...
- C++中GB2312字符串和UTF-8之间的转换
在编程过程中需要对字符串进行不同的转换,特别是Gb2312和Utf-8直接的转换.在几个开源的魔兽私服中,很多都是老外开发的,而暴雪为了能 够兼容世界上的各个字符集也使用了UTF-8.在中国使用VS( ...
随机推荐
- 把普通的JavaProject变成MavenProject
1,安装Maven Eclipse插件 2,在项目上点右键->Configure->Convert to maven project 3,在项目上点右键->Run as->Ru ...
- java正则表达式学习
1.简单认识正则: public class Test { public static void main(String[] args) { //简单认识正则 p("abc".ma ...
- python appium增加方法
1.测试过程中发现python client没有拨打电话的方法,因此去添加该方法 1.1查看源码 appium-base-driver/blob/master/lib/protocol/routes. ...
- Shell的for和select
环境准备: [root@nodchen-db01-test day07]# mkdir -p /server/scripts/day07/test/{test.txt,oldboy.txt,oldgi ...
- centos 7.5 安装mongodb
MongoDB安装和启动 从官网下载最新对应的版本然后解压,本文以3.6.9为例,将文件拷贝到opt目录下,然后解压: [root@localhost opt]# tar zxvf mongodb-l ...
- Flex 学习
Flex案例一: <html> <head> <meta http-equiv="Content-Type" content="text/h ...
- php使用inotify实现队列处理
php使用inotify实现队列处理参考如下文章:http://blog.jiunile.com/php%E4%BD%BF%E7%94%A8inotify%E5%AE%9E%E7%8E%B0%E9%9 ...
- Ubuntu14.04下hadoop-2.6.0单机配置和伪分布式配置
需要重新编译的教程:http://blog.csdn.net/ggz631047367/article/details/42460589 在Ubuntu下创建hadoop用户组和用户 hadoop的管 ...
- HttpClient上传下载文件
HttpClient上传下载文件 java HttpClient Maven依赖 <dependency> <groupId>org.apache.httpcomponents ...
- 如何分析 WindowsDump:Dump 起源与初始设置
https://www.qcloud.com/community/article/511817 转者注:让我感觉以前看蓝屏都白看了~~~原来蓝屏也可以分析具体原因. 适用场景:Windows 系列系统 ...
