orthodb
1、数据库

orthodb数据:
odb10v0_levels.tab.gz: NCBI taxonomy nodes where Ortho DB orthologous groups (OGs) are calculated
odb10v0_species.tab.gz: Ortho DB individual organism (aka species) ids based on NCBI taxonomy ids (mostly species level)
odb10v0_level2species.tab.gz: correspondence between level ids and species ids
odb10v0_genes.tab.gz: Ortho DB genes with some info
odb10v0_OGs.tab.gz: Ortho DB orthologous groups
odb10v0_OG2genes.tab.gz: OGs to genes correspondence
odb10v0_OG_xrefs.tab.gz: OG associations with GO, COG and InterPro ids
v9_v10_OGs_map.tab.gz mappings between the previous and current release orthologous group ids
odb10v0_fasta_<root>.tgz tar-ball with one fasta file per taxon id in the given root (bacteria,metazoa,fungi,plants)
2、odb10v0_levels.tab:
1. level NCBI taxonomy id
2. scientific name
3. total non-redundant count of genes in all underneath clustered species(在聚集的物种下面的所有的基因的总非重复计数)
4. total count of OGs built on it
5. total non-redundant count of species underneath

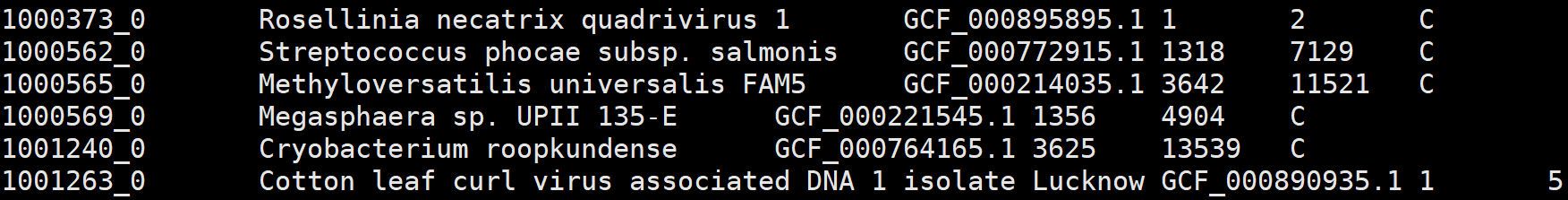
3、odb10v0_species.tab.gz
1. Ortho DB individual organism id, based on NCBI tax id
2. scientific name inherited from the most relevant NCBI tax id
3. genome asssembly id, when available
4. total count of clustered genes in this species
5. total count of the OGs it participates
6. mapping type, clustered(C) or mapped(M)
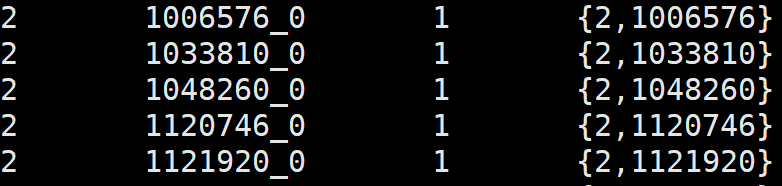
4、odb10v0_level2species.tab
1. top-most level NCBI tax id, one of [2,2157,2759,10239]
2. Ortho DB organism id
3. number of hops between the top-most level id and the NCBI tax id assiciated with the organism
4. ordered list of Ortho DB selected intermediate levels from the top-most level to the bottom one
5、odb10v0_genes.tab
1. Ortho DB unique gene id (not stable between releases)
2. organism tax id
3. protein original sequence id, as downloaded along with the sequence
4. Uniprot id, evaluated by mapping
5. ENSEMBL gene name, evaluated by mapping
6. NCBI gid, evaluated by mapping
7. description, evaluated by mapping
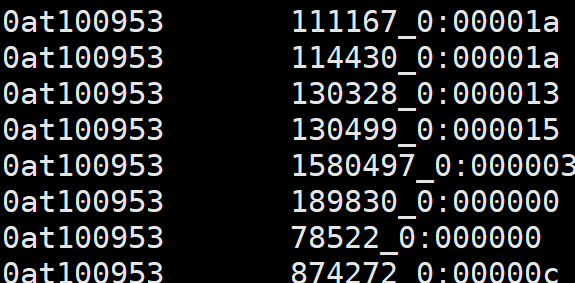
6、odb10v0_OG2genes.tab
1. OG unique id
2. Ortho DB gene id
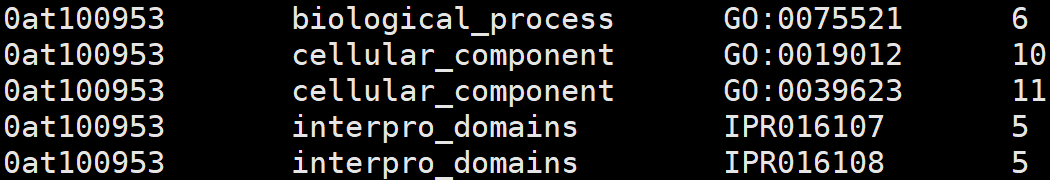
7、odb10v0_OG_xrefs.tab
1. OG unique id
2. external DB or DB section
3. external identifier
4. number of genes in the OG associated with the identifier
参考
https://www.orthodb.org/?page=filelist
orthodb的更多相关文章
- 【基因组预测】braker2基因结构注释要点记录
目录 流程使用 问题 记录下braker2的使用要点,以备忘记. 流程使用 braker2有很多流程,根据你的数据:组装的基因组.转录组.蛋白(同源,包括近缘或远缘)选择不同流程,官网有说明: htt ...
随机推荐
- 运行vbs脚本
VBS是基于Visual Basic的脚本语言. VBS的全称是:Microsoft Visual Basic Script Edition.(微软公司可视化BASIC脚本版). 其语言类似Visua ...
- java 怎样向一个已存在的文件中添加内容
如果想向某个文件最后添加内容,可使用FileWriter fw = new FileWriter("log.txt",true);在创建FileWriter时加个true就可以了. ...
- 20165205 2017-2018-2 《Java程序设计》第九周学习总结
20165205 2017-2018-2 <Java程序设计>第九周学习总结 教材学习内容总结 掌握URL类的使用方法 URL类的构造方法: public URL(String spec) ...
- gentoo: startx: drmsetmaster failed: permission denied
今天更新了 xorg-server 之后, startx 就进不了 X了,但是可以用 sudo startx 进入 X,所以感觉很奇怪. 后来终于在 gentoo 官方论坛上面找到答案了. https ...
- windows server 域分发与分配软件
参考网站:https://blog.csdn.net/southwind0/article/details/80734508 1.win 2008创建域 https://jingyan.baidu.c ...
- Linux——JDK配置
一.安装jdk-7u21-linux-x64.rpm文件 [root@centos6 local]# rpm –ivh jdk-7u21-linux-x64.rpm 二.防火墙开放8080端口 (在 ...
- otter 数据同步
阿里巴巴分布式数据库同步系统(解决中美异地机房) 基本介绍: https://github.com/alibaba/otter 快速使用: https://github.com/alibaba/ott ...
- 10.Linux 管道
简单地说,一个通道接受一个工具软件的输出,然后把那个输出输入到其它工具软件.使用UNIX/Linux的词汇,这个通道接受了一个过程的标准输出,并把这个标准的输出作为另一个过程的标准输入.如果你没有重新 ...
- Set和Map数据
es6新的数据结构 1.Set:构造函数 const s = new Set ([1,2,3]); console.log(s)//Set(3){1,2,3};[...s];//[1,2,3]cons ...
- Centos 7上安装Elasticsearch
1. 先安装jdk yum search java|grep jdk查看yum库中都有哪些jdk版本 yum install java-1.8.0-openjdk.x86_64 两次y确认 2. ce ...
