吴裕雄--天生自然 R语言开发学习:图形初阶(续一)





















# ----------------------------------------------------#
# R in Action (2nd ed): Chapter 3 #
# Getting started with graphs #
# requires that the Hmisc and RColorBrewer packages #
# have been installed #
# install.packages(c("Hmisc", "RColorBrewer")) #
#-----------------------------------------------------# par(ask=TRUE)
opar <- par(no.readonly=TRUE) # make a copy of current settings attach(mtcars) # be sure to execute this line plot(wt, mpg)
abline(lm(mpg~wt))
title("Regression of MPG on Weight")
# Input data for drug example
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40) plot(dose, drugA, type="b") opar <- par(no.readonly=TRUE) # make a copy of current settings
par(lty=2, pch=17) # change line type and symbol
plot(dose, drugA, type="b") # generate a plot
par(opar) # restore the original settings plot(dose, drugA, type="b", lty=3, lwd=3, pch=15, cex=2) # choosing colors
library(RColorBrewer)
n <- 7
mycolors <- brewer.pal(n, "Set1")
barplot(rep(1,n), col=mycolors) n <- 10
mycolors <- rainbow(n)
pie(rep(1, n), labels=mycolors, col=mycolors)
mygrays <- gray(0:n/n)
pie(rep(1, n), labels=mygrays, col=mygrays) # Listing 3.1 - Using graphical parameters to control graph appearance
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40)
opar <- par(no.readonly=TRUE)
par(pin=c(2, 3))
par(lwd=2, cex=1.5)
par(cex.axis=.75, font.axis=3)
plot(dose, drugA, type="b", pch=19, lty=2, col="red")
plot(dose, drugB, type="b", pch=23, lty=6, col="blue", bg="green")
par(opar) # Adding text, lines, and symbols
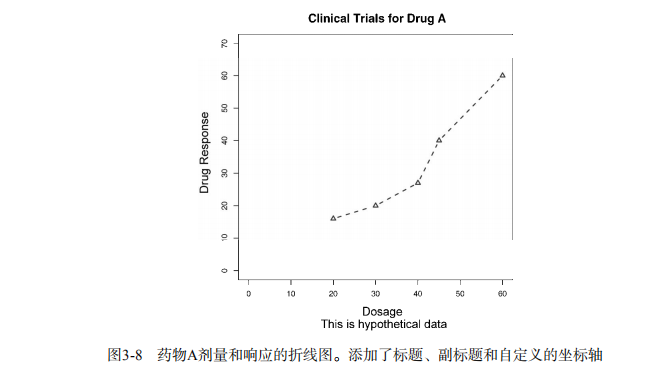
plot(dose, drugA, type="b",
col="red", lty=2, pch=2, lwd=2,
main="Clinical Trials for Drug A",
sub="This is hypothetical data",
xlab="Dosage", ylab="Drug Response",
xlim=c(0, 60), ylim=c(0, 70)) # Listing 3.2 - An Example of Custom Axes
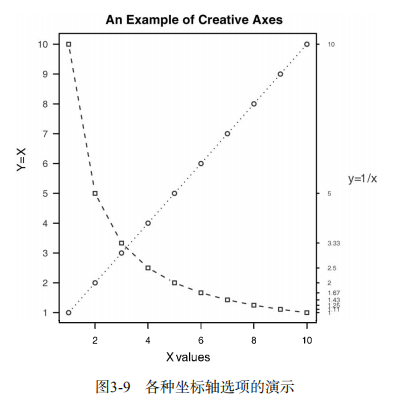
x <- c(1:10)
y <- x
z <- 10/x
opar <- par(no.readonly=TRUE)
par(mar=c(5, 4, 4, 8) + 0.1)
plot(x, y, type="b",
pch=21, col="red",
yaxt="n", lty=3, ann=FALSE)
lines(x, z, type="b", pch=22, col="blue", lty=2)
axis(2, at=x, labels=x, col.axis="red", las=2)
axis(4, at=z, labels=round(z, digits=2),
col.axis="blue", las=2, cex.axis=0.7, tck=-.01)
mtext("y=1/x", side=4, line=3, cex.lab=1, las=2, col="blue")
title("An Example of Creative Axes",
xlab="X values",
ylab="Y=X")
par(opar) # Listing 3.3 - Comparing Drug A and Drug B response by dose
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40)
opar <- par(no.readonly=TRUE)
par(lwd=2, cex=1.5, font.lab=2)
plot(dose, drugA, type="b",
pch=15, lty=1, col="red", ylim=c(0, 60),
main="Drug A vs. Drug B",
xlab="Drug Dosage", ylab="Drug Response")
lines(dose, drugB, type="b",
pch=17, lty=2, col="blue")
abline(h=c(30), lwd=1.5, lty=2, col="gray")
library(Hmisc)
minor.tick(nx=3, ny=3, tick.ratio=0.5)
legend("topleft", inset=.05, title="Drug Type", c("A","B"),
lty=c(1, 2), pch=c(15, 17), col=c("red", "blue"))
par(opar) # Example of labeling points
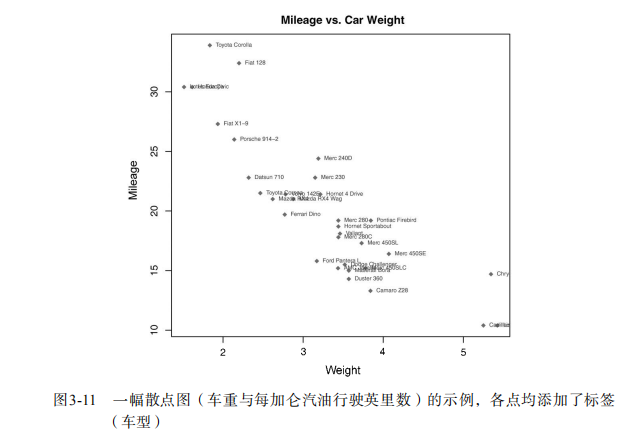
attach(mtcars)
plot(wt, mpg,
main="Mileage vs. Car Weight",
xlab="Weight", ylab="Mileage",
pch=18, col="blue")
text(wt, mpg,
row.names(mtcars),
cex=0.6, pos=4, col="red")
detach(mtcars) # View font families
opar <- par(no.readonly=TRUE)
par(cex=1.5)
plot(1:7,1:7,type="n")
text(3,3,"Example of default text")
text(4,4,family="mono","Example of mono-spaced text")
text(5,5,family="serif","Example of serif text")
par(opar) # Combining graphs
attach(mtcars)
opar <- par(no.readonly=TRUE)
par(mfrow=c(2,2))
plot(wt,mpg, main="Scatterplot of wt vs. mpg")
plot(wt,disp, main="Scatterplot of wt vs. disp")
hist(wt, main="Histogram of wt")
boxplot(wt, main="Boxplot of wt")
par(opar)
detach(mtcars) attach(mtcars)
opar <- par(no.readonly=TRUE)
par(mfrow=c(3,1))
hist(wt)
hist(mpg)
hist(disp)
par(opar)
detach(mtcars) attach(mtcars)
layout(matrix(c(1,1,2,3), 2, 2, byrow = TRUE))
hist(wt)
hist(mpg)
hist(disp)
detach(mtcars) attach(mtcars)
layout(matrix(c(1, 1, 2, 3), 2, 2, byrow = TRUE),
widths=c(3, 1), heights=c(1, 2))
hist(wt)
hist(mpg)
hist(disp)
detach(mtcars) # Listing 3.4 - Fine placement of figures in a graph
opar <- par(no.readonly=TRUE)
par(fig=c(0, 0.8, 0, 0.8))
plot(mtcars$mpg, mtcars$wt,
xlab="Miles Per Gallon",
ylab="Car Weight")
par(fig=c(0, 0.8, 0.55, 1), new=TRUE)
boxplot(mtcars$mpg, horizontal=TRUE, axes=FALSE)
par(fig=c(0.65, 1, 0, 0.8), new=TRUE)
boxplot(mtcars$wt, axes=FALSE)
mtext("Enhanced Scatterplot", side=3, outer=TRUE, line=-3)
par(opar)
吴裕雄--天生自然 R语言开发学习:图形初阶(续一)的更多相关文章
- 吴裕雄--天生自然 R语言开发学习:聚类分析(续一)
#-------------------------------------------------------# # R in Action (2nd ed): Chapter 16 # # Clu ...
- 吴裕雄--天生自然 R语言开发学习:时间序列(续三)
#-----------------------------------------# # R in Action (2nd ed): Chapter 15 # # Time series # # r ...
- 吴裕雄--天生自然 R语言开发学习:时间序列(续二)
#-----------------------------------------# # R in Action (2nd ed): Chapter 15 # # Time series # # r ...
- 吴裕雄--天生自然 R语言开发学习:时间序列(续一)
#-----------------------------------------# # R in Action (2nd ed): Chapter 15 # # Time series # # r ...
- 吴裕雄--天生自然 R语言开发学习:方差分析(续二)
#-------------------------------------------------------------------# # R in Action (2nd ed): Chapte ...
- 吴裕雄--天生自然 R语言开发学习:方差分析(续一)
#-------------------------------------------------------------------# # R in Action (2nd ed): Chapte ...
- 吴裕雄--天生自然 R语言开发学习:回归(续四)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
- 吴裕雄--天生自然 R语言开发学习:回归(续三)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
- 吴裕雄--天生自然 R语言开发学习:回归(续二)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
- 吴裕雄--天生自然 R语言开发学习:回归(续一)
#------------------------------------------------------------# # R in Action (2nd ed): Chapter 8 # # ...
随机推荐
- 前端Json 增加,删除,修改元素(包含json数组处理)
一:基础JSON对象 二:JSON数组数据 以下为增删修改方法: <!DOCTYPE html> <html lang="en"> <head> ...
- c#学习笔记05——数组&集合
数组 声明数组 .一维数组的定义: 数据类型[] 数组名=new 数据类型[大小]; eg: ]; ,,,,}; ]; .多维数组的定义 ,];//定义二维数组 ,,];//定义三维数组 多维数组可以 ...
- linux服务器CentOS7安装node.js
方维一元夺宝2.0版本,很多用户面临机器人自动执行任务.采集计划一直无法开启的问题. 这个需要开启node.js分享给大家. 1.获取node.js资源 V5.x: curl --silent --l ...
- 致 Python 初学者们!
前言 在 Python 进阶的过程中,相信很多同学应该大致上学习了很多 Python 的基础知识,也正在努力成长.在此期间,一定遇到了很多的困惑,对未来的学习方向感到迷茫.我非常理解你们所面临的处 ...
- RL78 定义常量变量在指定的地址方法
若想定义的常量地址在远端寻址,定义section段时 如定义MCU_INFOR段 则段名为MCU_INFOR_f 后缀需要添加f,近端寻址添加n. 程序中定义常量 需要使用#pragma 指 ...
- DFS---迷宫问题
#include<iostream> #include<string> #include<cstring> using namespace std;//dfs in ...
- 用户界面编程模式 MVC MVP MVVM
用户界面编程模式 MVC MVP MVVM 程序 = 数据 + 算法 数据:就是待处理的东西 算法:就是代码 涉及到人机交互的程序,不可避免涉及到界面和界面上显示的数据原始方式是界面代码和逻辑代码糅合 ...
- liquibase 注意事项
liquibase 一个changelog中有多个sql语句时,如果后边报错,前边的sql执行成功后是不会回滚的,所以最好分开写sql <changeSet author="lihao ...
- 安卓ButtomBar实现方法
这里ButtomBar有3个items,分别有icon和文字,在当前fragment时,所属的icon和文字会显示不同颜色. 1. 首先要准好ICON素材,命名规范要清楚. 2. 实现这个Buttom ...
- Codeforces Round #576 (Div. 2) D. Welfare State
http://codeforces.com/contest/1199/problem/D Examples input1 output1 input2 output2 Note In the firs ...
