[占位-未完成]scikit-learn一般实例之十二:用于RBF核的显式特征映射逼近
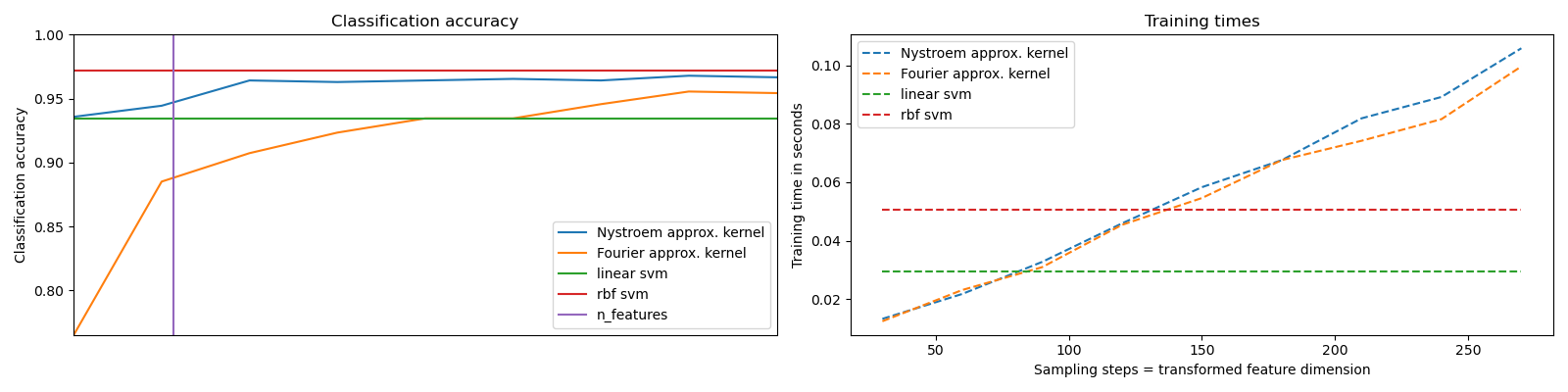
It shows how to use RBFSampler and Nystroem to approximate the feature map of an RBF kernel for classification with an SVM on the digits dataset. Results using a linear SVM in the original space, a linear SVM using the approximate mappings and using a kernelized SVM are compared. Timings and accuracy for varying amounts of Monte Carlo samplings (in the case of RBFSampler, which uses random Fourier features) and different sized subsets of the training set (for Nystroem) for the approximate mapping are shown.
Please note that the dataset here is not large enough to show the benefits of kernel approximation, as the exact SVM is still reasonably fast.
Sampling more dimensions clearly leads to better classification results, but comes at a greater cost. This means there is a tradeoff between runtime and accuracy, given by the parameter n_components. Note that solving the Linear SVM and also the approximate kernel SVM could be greatly accelerated by using stochastic gradient descent via sklearn.linear_model.SGDClassifier. This is not easily possible for the case of the kernelized SVM.
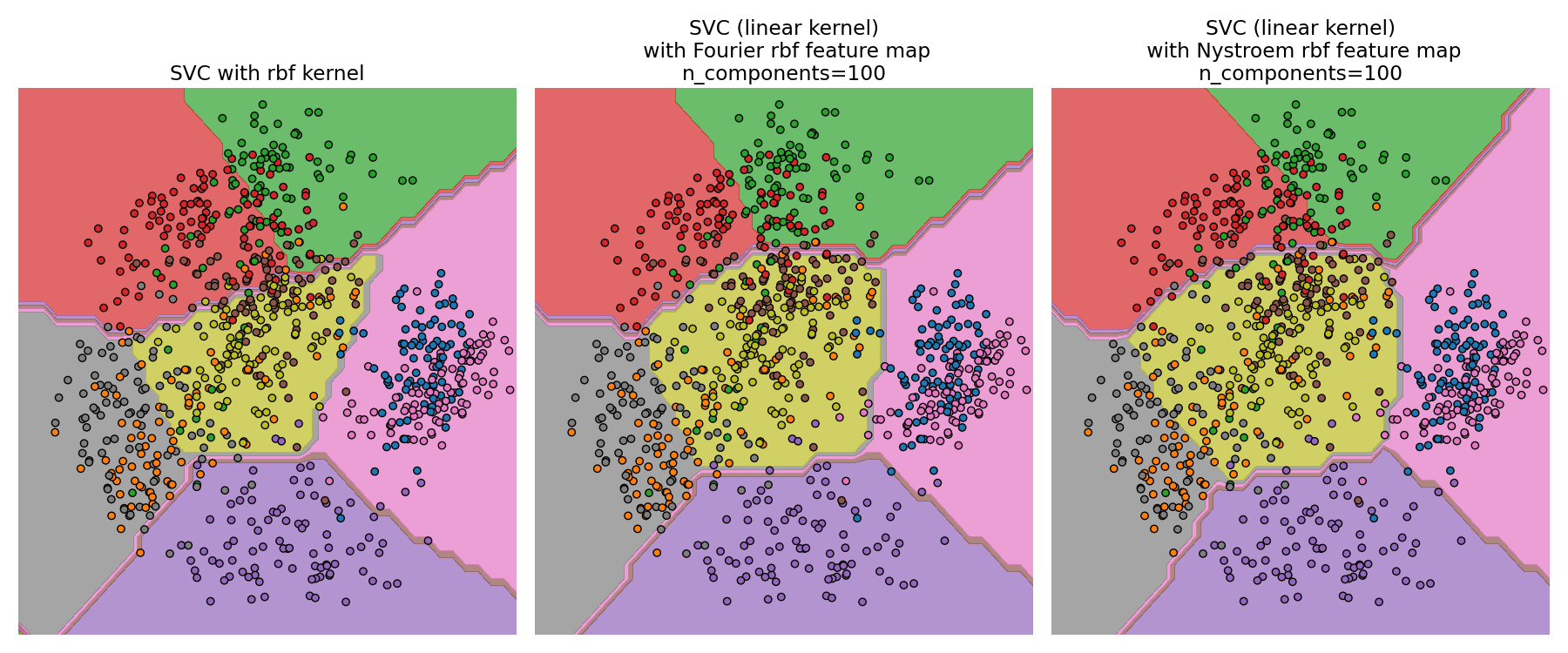
The second plot visualized the decision surfaces of the RBF kernel SVM and the linear SVM with approximate kernel maps. The plot shows decision surfaces of the classifiers projected onto the first two principal components of the data. This visualization should be taken with a grain of salt since it is just an interesting slice through the decision surface in 64 dimensions. In particular note that a datapoint (represented as a dot) does not necessarily be classified into the region it is lying in, since it will not lie on the plane that the first two principal components span.
The usage of RBFSampler and Nystroem is described in detail in Kernel Approximation.
print(__doc__)
# Author: Gael Varoquaux <gael dot varoquaux at normalesup dot org>
# Andreas Mueller <amueller@ais.uni-bonn.de>
# License: BSD 3 clause
# Standard scientific Python imports
import matplotlib.pyplot as plt
import numpy as np
from time import time
# Import datasets, classifiers and performance metrics
from sklearn import datasets, svm, pipeline
from sklearn.kernel_approximation import (RBFSampler,
Nystroem)
from sklearn.decomposition import PCA
# The digits dataset
digits = datasets.load_digits(n_class=9)
# To apply an classifier on this data, we need to flatten the image, to
# turn the data in a (samples, feature) matrix:
n_samples = len(digits.data)
data = digits.data / 16.
data -= data.mean(axis=0)
# We learn the digits on the first half of the digits
data_train, targets_train = data[:n_samples / 2], digits.target[:n_samples / 2]
# Now predict the value of the digit on the second half:
data_test, targets_test = data[n_samples / 2:], digits.target[n_samples / 2:]
#data_test = scaler.transform(data_test)
# Create a classifier: a support vector classifier
kernel_svm = svm.SVC(gamma=.2)
linear_svm = svm.LinearSVC()
# create pipeline from kernel approximation
# and linear svm
feature_map_fourier = RBFSampler(gamma=.2, random_state=1)
feature_map_nystroem = Nystroem(gamma=.2, random_state=1)
fourier_approx_svm = pipeline.Pipeline([("feature_map", feature_map_fourier),
("svm", svm.LinearSVC())])
nystroem_approx_svm = pipeline.Pipeline([("feature_map", feature_map_nystroem),
("svm", svm.LinearSVC())])
# fit and predict using linear and kernel svm:
kernel_svm_time = time()
kernel_svm.fit(data_train, targets_train)
kernel_svm_score = kernel_svm.score(data_test, targets_test)
kernel_svm_time = time() - kernel_svm_time
linear_svm_time = time()
linear_svm.fit(data_train, targets_train)
linear_svm_score = linear_svm.score(data_test, targets_test)
linear_svm_time = time() - linear_svm_time
sample_sizes = 30 * np.arange(1, 10)
fourier_scores = []
nystroem_scores = []
fourier_times = []
nystroem_times = []
for D in sample_sizes:
fourier_approx_svm.set_params(feature_map__n_components=D)
nystroem_approx_svm.set_params(feature_map__n_components=D)
start = time()
nystroem_approx_svm.fit(data_train, targets_train)
nystroem_times.append(time() - start)
start = time()
fourier_approx_svm.fit(data_train, targets_train)
fourier_times.append(time() - start)
fourier_score = fourier_approx_svm.score(data_test, targets_test)
nystroem_score = nystroem_approx_svm.score(data_test, targets_test)
nystroem_scores.append(nystroem_score)
fourier_scores.append(fourier_score)
# plot the results:
plt.figure(figsize=(8, 8))
accuracy = plt.subplot(211)
# second y axis for timeings
timescale = plt.subplot(212)
accuracy.plot(sample_sizes, nystroem_scores, label="Nystroem approx. kernel")
timescale.plot(sample_sizes, nystroem_times, '--',
label='Nystroem approx. kernel')
accuracy.plot(sample_sizes, fourier_scores, label="Fourier approx. kernel")
timescale.plot(sample_sizes, fourier_times, '--',
label='Fourier approx. kernel')
# horizontal lines for exact rbf and linear kernels:
accuracy.plot([sample_sizes[0], sample_sizes[-1]],
[linear_svm_score, linear_svm_score], label="linear svm")
timescale.plot([sample_sizes[0], sample_sizes[-1]],
[linear_svm_time, linear_svm_time], '--', label='linear svm')
accuracy.plot([sample_sizes[0], sample_sizes[-1]],
[kernel_svm_score, kernel_svm_score], label="rbf svm")
timescale.plot([sample_sizes[0], sample_sizes[-1]],
[kernel_svm_time, kernel_svm_time], '--', label='rbf svm')
# vertical line for dataset dimensionality = 64
accuracy.plot([64, 64], [0.7, 1], label="n_features")
# legends and labels
accuracy.set_title("Classification accuracy")
timescale.set_title("Training times")
accuracy.set_xlim(sample_sizes[0], sample_sizes[-1])
accuracy.set_xticks(())
accuracy.set_ylim(np.min(fourier_scores), 1)
timescale.set_xlabel("Sampling steps = transformed feature dimension")
accuracy.set_ylabel("Classification accuracy")
timescale.set_ylabel("Training time in seconds")
accuracy.legend(loc='best')
timescale.legend(loc='best')
# visualize the decision surface, projected down to the first
# two principal components of the dataset
pca = PCA(n_components=8).fit(data_train)
X = pca.transform(data_train)
# Generate grid along first two principal components
multiples = np.arange(-2, 2, 0.1)
# steps along first component
first = multiples[:, np.newaxis] * pca.components_[0, :]
# steps along second component
second = multiples[:, np.newaxis] * pca.components_[1, :]
# combine
grid = first[np.newaxis, :, :] + second[:, np.newaxis, :]
flat_grid = grid.reshape(-1, data.shape[1])
# title for the plots
titles = ['SVC with rbf kernel',
'SVC (linear kernel)\n with Fourier rbf feature map\n'
'n_components=100',
'SVC (linear kernel)\n with Nystroem rbf feature map\n'
'n_components=100']
plt.tight_layout()
plt.figure(figsize=(12, 5))
# predict and plot
for i, clf in enumerate((kernel_svm, nystroem_approx_svm,
fourier_approx_svm)):
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
plt.subplot(1, 3, i + 1)
Z = clf.predict(flat_grid)
# Put the result into a color plot
Z = Z.reshape(grid.shape[:-1])
plt.contourf(multiples, multiples, Z, cmap=plt.cm.Paired)
plt.axis('off')
# Plot also the training points
plt.scatter(X[:, 0], X[:, 1], c=targets_train, cmap=plt.cm.Paired)
plt.title(titles[i])
plt.tight_layout()
plt.show()
[占位-未完成]scikit-learn一般实例之十二:用于RBF核的显式特征映射逼近的更多相关文章
- C++面向对象类的实例题目十二
题目描述: 写一个程序计算正方体.球体和圆柱体的表面积和体积 程序代码: #include<iostream> #define PAI 3.1415 using namespace std ...
- 数据可视化实例(十二): 发散型条形图 (matplotlib,pandas)
https://datawhalechina.github.io/pms50/#/chapter10/chapter10 如果您想根据单个指标查看项目的变化情况,并可视化此差异的顺序和数量,那么散型条 ...
- Java开发笔记(七十二)Java8新增的流式处理
通过前面几篇文章的学习,大家应能掌握几种容器类型的常见用法,对于简单的增删改和遍历操作,各容器实例都提供了相应的处理方法,对于实际开发中频繁使用的清单List,还能利用Arrays工具的asList方 ...
- 框架源码系列十二:Mybatis源码之手写Mybatis
一.需求分析 1.Mybatis是什么? 一个半自动化的orm框架(Object Relation Mapping). 2.Mybatis完成什么工作? 在面向对象编程中,我们操作的都是对象,Myba ...
- [占位-未完成]scikit-learn一般实例之十:核岭回归和SVR的比较
[占位-未完成]scikit-learn一般实例之十:核岭回归和SVR的比较
- [占位-未完成]scikit-learn一般实例之十一:异构数据源的特征联合
[占位-未完成]scikit-learn一般实例之十一:异构数据源的特征联合 Datasets can often contain components of that require differe ...
- scikit learn 模块 调参 pipeline+girdsearch 数据举例:文档分类 (python代码)
scikit learn 模块 调参 pipeline+girdsearch 数据举例:文档分类数据集 fetch_20newsgroups #-*- coding: UTF-8 -*- import ...
- Scikit Learn: 在python中机器学习
转自:http://my.oschina.net/u/175377/blog/84420#OSC_h2_23 Scikit Learn: 在python中机器学习 Warning 警告:有些没能理解的 ...
- 《Android群英传》读书笔记 (5) 第十一章 搭建云端服务器 + 第十二章 Android 5.X新特性详解 + 第十三章 Android实例提高
第十一章 搭建云端服务器 该章主要介绍了移动后端服务的概念以及Bmob的使用,比较简单,所以略过不总结. 第十三章 Android实例提高 该章主要介绍了拼图游戏和2048的小项目实例,主要是代码,所 ...
随机推荐
- 【小计】新人Tostring前忘记Null判断的处理
ToString和string.Concat(可屏蔽Null的异常)性能相差不大,一些中小项目完全可以用Concat(新人容易忘记判断Null的情况,遇到太多了,所以建议重写tostring方法,内部 ...
- ASP.NET MVC5+EF6+EasyUI 后台管理系统(69)-微信公众平台开发-功能概述
系列目录 为什么要先发这个文章? 因为接下来的文章是关于微信开发的系列,心中一定要有一个概念,知道自己接下来要做什么功能. 而且微信到处都是坑,我首先要把微信与本地跑通起来才敢发布,否则中间出现坑导致 ...
- 基于netty http协议栈的轻量级流程控制组件的实现
今儿个是冬至,所谓“冬大过年”,公司也应景五点钟就放大伙儿回家吃饺子喝羊肉汤了,而我本着极高的职业素养依然坚持留在公司(实则因为没饺子吃没羊肉汤喝,只能呆公司吃食堂……).趁着这一个多小时的时间,想跟 ...
- python+uwsgi导致redis无法长链接引起性能下降问题记录
今天在部署python代码到预生产环境时,web站老是出现redis链接未初始化,无法连接到服务的提示,比对了一下开发环境与测试环境代码,完全一致,然后就是查看各种日志,排查了半天也没有查明是什么原因 ...
- ZKWeb网页框架1.2正式发布
发行日志 https://github.com/zkweb-framework/ZKWeb/blob/master/ReleaseNotes/ReleaseNote.1.2.md 主要改动 更新 ZK ...
- webpack学习总结
前言 在还未接触webpack,就有几个疑问: 1. webpack本质上是什么? 2. 跟异步模块加载有关系吗? 3. 可否生成多个文件,一定是一个? 4. 被引用的文件有其他异步加载模块怎么办? ...
- 第10章 Shell编程(4)_流程控制
5. 流程控制 5.1 if语句 (1)格式: 格式1 格式2 多分支if if [ 条件判断式 ];then #程序 else #程序 fi if [ 条件判断式 ] then #程序 else # ...
- 性能测试工具 wrk 安装与使用
介绍 今天给大家介绍一款开源的性能测试工具 wrk,简单易用,没有Load Runner那么复杂,他和 apache benchmark(ab)同属于性能测试工具,但是比 ab 功能更加强大,并且可以 ...
- AutoMapper(一)
返回总目录 映射前后操作 偶尔有时候,在映射发生之前或之后,你可能需要执行一些自定义的逻辑.这可能是很少见的事情,因为在AutoMapper之外处理这些事情是更明显的.你可以创建一个映射前后的全局操作 ...
- 使用mapreduce计算环比的实例
最近做了一个小的mapreduce程序,主要目的是计算环比值最高的前5名,本来打算使用spark计算,可是本人目前spark还只是简单看了下,因此就先改用mapreduce计算了,今天和大家分享下这个 ...
