edgeR
1)简介
edgeR作用对象是count文件,rows 代表基因,行代表文库,count代表的是比对到每个基因的reads数目。它主要关注的是差异表达分析,而不是定量基因表达水平。
edgeR works on a table of integer read counts, with rows corresponding to genes and columns to independent libraries. The counts represent the total number of reads aligning to each gene (or other genomic locus).edgeR is concerned with differential expression analysis rather than with the quantification of expression levels. It is concerned with relative changes in expression levels between conditions,but not directly with estimating absolute expression levels.
edgeR作用的是真实的比对统计,因此不建议用预测的转录本
Note that edgeR is designed to work with actual read counts. We not recommend that predicted transcript abundances are input the edgeR in place of actual counts.
归一化原因:
技术原因影响差异表达分析:
1)Sequencing depth:统计测序深度(即代表的是library size);
2)RNA composition:个别异常高表达基因导致其它基因采样不足
3)GC content: sample-specific effects for GC-content can be detected
4)sample-specific effects for gene length have been detected
注意:edgeR必须是原始表达量,而不能是rpkm等矫正过的。
Note that normalization in edgeR is model-based, and the original read counts are not themselves transformed. This means that users should not transform the read counts in any way before inputing them to edgeR. For example, users should not enter RPKM or FPKM values to edgeR in place of read counts. Such quantities will prevent edgeR from correctly estimating the mean-variance relationship in the data, which is a crucial to the statistical strategies underlying edgeR.Similarly, users should not add artificial values to the counts before inputing them to edgeR.
2)安装
if("edgeR" %in% rownames(installed.packages()) == FALSE) {source("http://bioconductor.org/biocLite.R");biocLite("edgeR")}
suppressMessages(library(edgeR))
ls('package:edgeR')
3)矩阵构建及差异分析
需要构建2个矩阵:1、表达矩阵;2、分组矩阵( 实验设计);
-------------------------------------------------------表达矩阵-----------------------------------------
3.1、读取表达矩阵文件(Reading in the data)
#读取文件
rawdata <- read.delim("E:/software/R/R-3.5.0/library/edgeR/Meta/TableS1.txt", check.names=FALSE, stringsAsFactors=FALSE)
head(rawdata)
3.2 、构建DGEList对象
这里因为已经有rawdata的count文件,因此直接用DGEList()函数就行了,否则要用readDGE()函数
y <- DGEList(counts=rawdata[,4:9], genes=rawdata[,1:3])##构建DGEList对象
DGEList对象主要有三部分:
1、counts矩阵:包含的是整数counts;
2、samples数据框:包含的是文库(sample)信息。包含 lib.size列 :for the library size (sequencing depth) for each sample,如果不自定义, the library sizes will be computed from the column sums of the counts。其中还有一个group列,用于指定每个sample组信息
3、一个可选的数据框genes:gene的注释信息
3.3)数据注释( Annotation)
这里主要是因为该文章数据是前好多年的,因此需要过滤,symbol更新等。
1)The study was undertaken a few years ago, so not all of the RefSeq IDs provided by match RefSeq IDs currently in use. We retain only those transcripts with IDs in the current NCBI annotation, which is provided by the org.HS.eg.db package
2)因为edgeR默认使用NCBI中refSeq的ID,所以通过refseq Id 找到entrezID,然后通过entrezID对symbol更新
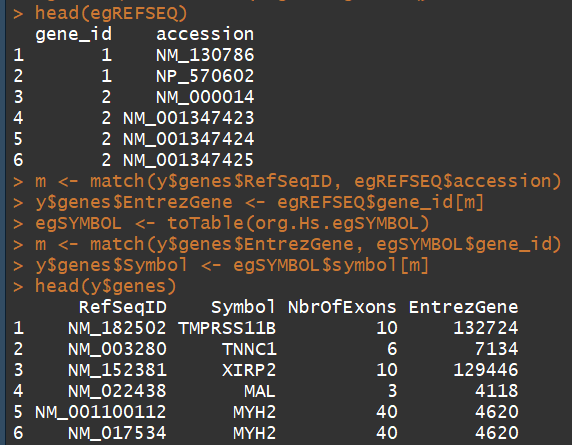
#######retain only those transcripts with IDs in the current NCBI annotation provided by the org.HS.eg.db######
library(org.Hs.eg.db)
idfound <- y$genes$RefSeqID %in% mappedRkeys(org.Hs.egREFSEQ)
y <- y[idfound,]
dim(y) ##15550 6
###################### 在注释中加入 Entrez Gene IDs #########################
egREFSEQ <- toTable(org.Hs.egREFSEQ)
m <- match(y$genes$RefSeqID, egREFSEQ$accession)
y$genes$EntrezGene <- egREFSEQ$gene_id[m]
#####################用Entrez Gene IDs更新gene symbols##########################
egSYMBOL <- toTable(org.Hs.egSYMBOL)
m <- match(y$genes$EntrezGene, egSYMBOL$gene_id)
y$genes$Symbol <- egSYMBOL$symbol[m]
head(y$genes)
3.4) 过滤和归一化(Filtering and normalization)
过滤一:Different RefSeq transcripts for the same gene symbol count predominantly the same reads. So we keep one transcript for each gene symbol. We choose the transcript with highest overall count:
o <- order(rowSums(y$counts), decreasing=TRUE)
y <- y[o,]
d <- duplicated(y$genes$Symbol)
y <- y[!d,]
nrow(y)
过滤二:Normally we would also filter lowly expressed genes.For this data, all transcripts already have at least 50 reads for all samples of at least one of the tissues types.
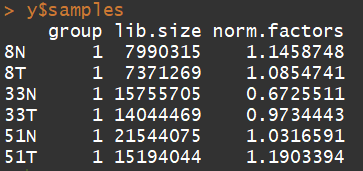
y$samples$lib.size <- colSums(y$counts) #Recompute the library sizes
###############################Use Entrez Gene IDs as row names:#####################
rownames(y$counts) <- rownames(y$genes) <- y$genes$EntrezGene
y$genes$EntrezGene <- NULL
归一化:TMM normalization is applied to this dataset to account for compositional difference between the libraries.
y <- calcNormFactors(y)
y$samples
3.5) 数据的探索(Data exploration)
样本间关系(samples for outliers and for other relationships)
plotMDS(y)
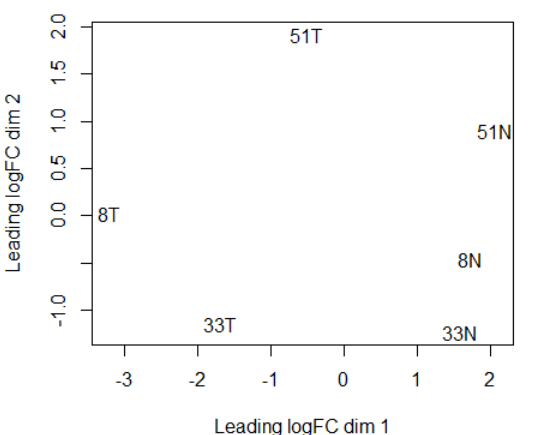
PC1将tumor和nomal组分开,PC2 大略和病号对应。也侧面体现了肿瘤组的异质性
--------------------------分组矩阵(根据实验设计、目的)--------------------------------
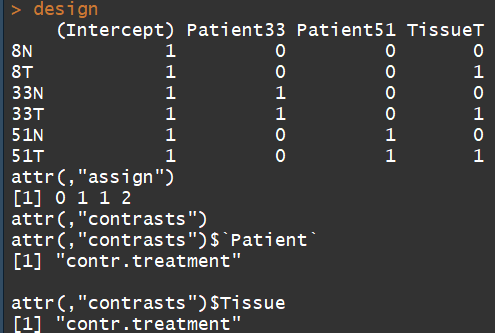
Here we want to test for differential expression between tumour and normal tissues within patients, i.e. adjusting for differences between patients.
Patient <- factor(c(8,8,33,33,51,51))
Tissue <- factor(c("N","T","N","T","N","T"))
data.frame(Sample=colnames(y),Patient,Tissue)
design <- model.matrix(~Patient+Tissue)
rownames(design) <- colnames(y)
design
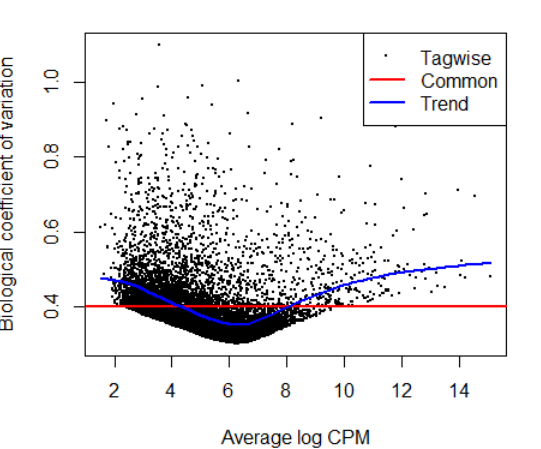
3.4)Estimating the dispersion(estimate the NB dispersion for the dataset.)
y <- estimateDisp(y, design, robust=TRUE)
y$common.dispersion #0.1594505
plotBCV(y)
-----------------------------------差异分析-----------------------------------------
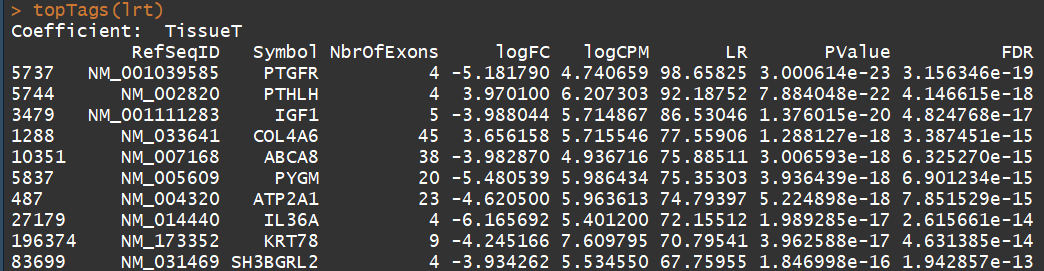
3.5) 差异分析(Differential expression)
fit <- glmFit(y, design)
lrt <- glmLRT(fit)
topTags(lrt)
summary(decideTests(lrt))
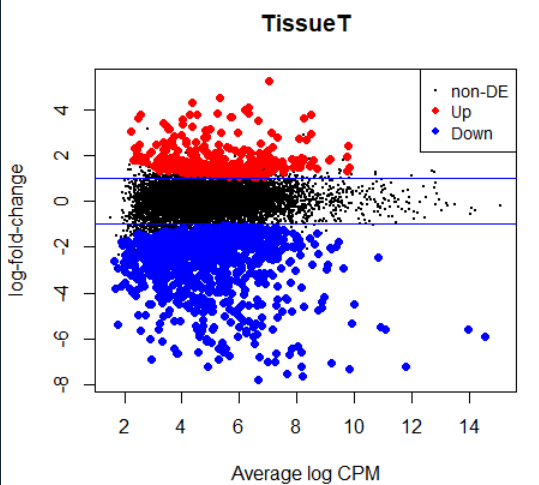
plotMD(lrt)
abline(h=c(-1, 1), col="blue")

------------------------------- Gene ontology analysis----------------------------------------
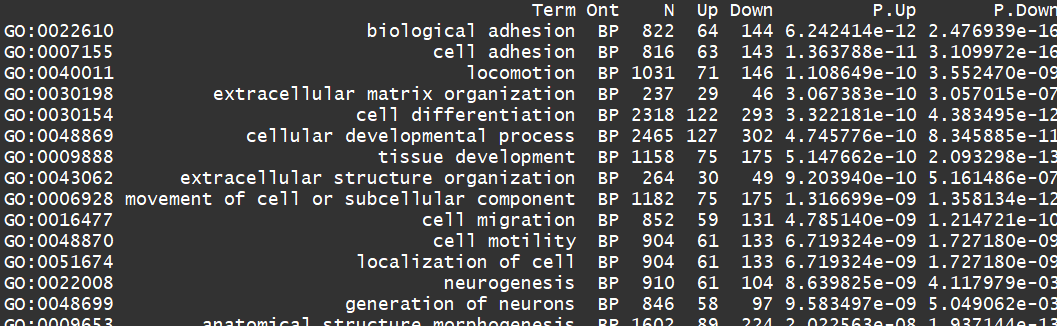
对上调的基因进行BP分析
go <- goana(lrt)
topGO(go, ont="BP", sort="Up", n=30)
edgeR的更多相关文章
- cuffdiff 和 edgeR 对差异表达基因的描述
ASE又走到了关键的一步 要生成能决定是否有差异表达的table. 准备借鉴一下cuffdiff和edgeR 的结果 cuffdiff对差异表达基因的描述: 一共十四列: 第一列, test_id ...
- 简单使用DESeq2/EdgeR做差异分析
简单使用DESeq2/EdgeR做差异分析 Posted: 五月 07, 2017 Under: Transcriptomics By Kai no Comments DESeq2和EdgeR都 ...
- 使用limma、Glimma和edgeR,RNA-seq数据分析易如反掌
使用limma.Glimma和edgeR,RNA-seq数据分析易如反掌 Charity Law1, Monther Alhamdoosh2, Shian Su3, Xueyi Dong3, Luyi ...
- edgeR使用学习【转载】
转自:http://yangl.net/2016/09/27/edger_usage/ 1.Quick start 2. 利用edgeR分析RNA-seq鉴别差异表达基因: #加载软件包 librar ...
- 用TCGA收集的mRNA表达数据作差异表达
做差异表达的软件DEseq和edgeR所需要的数据格式必须是原始counts,经过normalization和log2后的数据都不适合,所以对于做差异表达计算的童鞋可以使用ExperimentHub下 ...
- sql是最成功的第四代语言
SQL发展的前世今生 很多年前,两名年轻的IBM研究员将一门关系型语言带到了数据库领域,旨在使用声明性的方式来操作数据.从Don Chamberlin和Ramond Boyce发表"SEQU ...
- RNA-seq标准化
你的 heatmap 可能用错数据了 (组间表达量标准化) http://www.genek.tv/article/24 RNA-seq的标准化方法罗列 https://www.jianshu.com ...
- 史上最全 | 39个RNAseq分析工具与对比
文献:Sahraeian S M E, Mohiyuddin M, Sebra R, et al. Gaining comprehensive biological insight into the ...
- RNA-seq中的基因表达量计算和表达差异分析
RNA-seq中的基因表达量计算和表达差异分析 差异分析的步骤:1)比对:2) read count计算:3) read count的归一化:4)差异表达分析: 背景知识:1)比对:普通比对: BWA ...
随机推荐
- ubuntu初次设置root密码
初次安装ubuntu创建的用户不是root用户,但是需要root权限的时候又需要密码,那么如何设置密码呢? 很简单.如下几步操作
- jquery dataTable的学习
http://www.cnblogs.com/nier/archive/2012/03/19/2406450.html 分页 bPaginite:true;是否启用分页功能 sPaginationTy ...
- R语言学习——数据分析
complete.cases查看缺失值 箱图:
- 杂项:Office Visio
ylbtech-杂项:Office Visio Office Visio 是一款便于IT和商务人员就复杂信息.系统和流程进行可视化处理.分析和交流的软件.使用具有专业外观的 Office Visio ...
- 1067 Sort with Swap(0, i) (25 分)
1067 Sort with Swap(0, i) (25 分) Given any permutation of the numbers {0, 1, 2,..., N−1}, it is easy ...
- 递归神经网络(Recursive Neural Network, RNN)
信息往往还存在着诸如树结构.图结构等更复杂的结构.这就需要用到递归神经网络 (Recursive Neural Network, RNN),巧合的是递归神经网络的缩写和循环神经网络一样,也是RNN,递 ...
- 最简单的TCP、UDP案例及各函数的详细解释
TCP: server #include "stdafx.h" #include<iostream> #define BUF_SZIE 64 #include &quo ...
- FIO read测试结果偏离
工作中发现一个fio问题,测试组测试出来的数据read速度一个是17.0G/s,一个是13.2G/s.要知道我后台只有24块7.2k RPM的机械硬盘啊!怎么也不可能有这样的速度. 回家之后我模拟了实 ...
- XPath 常用语法札记
* 不包含属性的元素 例如不包含属性的span: span[not(@*)] * 文本包含某部分的元素 例如文本包含Rank的元素: *[contains(text(),'Rank')] * 选择匹配 ...
- Convolutional Neural Networks
卷积神经网络(Convolutional Neural Networks/ CNN/ConvNets) 卷积神经网络和普通神经网络十分相似: 组成它们的神经元都具有可学习的权重(weights)和偏置 ...
