杭电20题 Human Gene Functions
A human gene can be identified through a series of time-consuming biological experiments, often with the help of computer programs. Once a sequence of a gene is obtained, the next job is to determine its function. One of the methods for biologists to use in determining the function of a new gene sequence that they have just identified is to search a database with the new gene as a query. The database to be searched stores many gene sequences and their functions many researchers have been submitting their genes and functions to the database and the database is freely accessible through the Internet.
A database search will return a list of gene sequences from the database that are similar to the query gene. Biologists assume that sequence similarity often implies functional similarity. So, the function of the new gene might be one of the functions that the genes from the list have. To exactly determine which one is the right one another series of biological experiments will be needed.
Your job is to make a program that compares two genes and determines their similarity as explained below. Your program may be used as a part of the database search if you can provide an efficient one. Given two genes AGTGATG and GTTAG, how similar are they? One of the methods to measure the similarity of two genes is called alignment. In an alignment, spaces are inserted, if necessary, in appropriate positions of the genes to make them equally long and score the resulting genes according to a scoring matrix.
For example, one space is inserted into AGTGATG to result in AGTGAT-G, and three spaces are inserted into GTTAG to result in GT--TAG. A space is denoted by a minus sign (-). The two genes are now of equal length. These two strings are aligned:
AGTGAT-G -GT--TAG
In this alignment, there are four matches, namely, G in the second position, T in the third, T in the sixth, and G in the eighth. Each pair of aligned characters is assigned a score according to the following scoring matrix.
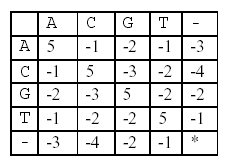 denotes that a space-space match is not allowed. The score of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9.
denotes that a space-space match is not allowed. The score of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9. Of course, many other alignments are possible. One is shown below (a different number of spaces are inserted into different positions):
AGTGATG -GTTA-G
This alignment gives a score of (-3)+5+5+(-2)+5+(-1) +5=14. So, this one is better than the previous one. As a matter of fact, this one is optimal since no other alignment can have a higher score. So, it is said that the similarity of the two genes is 14.
题意:匹配是 求最大的匹配和;可以加入空格;
求最大的匹配和;可以加入空格;
思路:思路实际上是求两个字符串的最大公共子序列的思路;
AC代码:
#include<iostream>
#include<cstdio>
#include<cstring>
#include<string>
#include<map> using namespace std; int max(int a,int b,int c)
{
return a>(b>c?b:c)?a:(b>c?b:c);
} int main()
{
freopen("1.txt","r",stdin);
int t;
string str1,str2;
int i,j;
int dp[][]={};
int s[][]={
{,-,-,-,-},
{-,,-,-,-},
{-,-,,-,-},
{-,-,-,,-},
{-,-,-,-,}};
map<char,int> k;
k['A']=;
k['C']=;
k['G']=;
k['T']=;
k['-']=;
cout<<k['A']<<endl<<k['C']<<endl<<k['G']<<endl<<k['T']<<endl<<k['-']<<endl;
cin>>t;
int n1,n2;
while(t)
{
memset(dp,,sizeof(dp));
cin>>n1>>str1>>n2>>str2;
for(i=;i<=n1;i++)
{
dp[i][]=dp[i-][]+s[k[str1[i-]]][k['-']];
}
for(i=;i<=n2;i++)
{
dp[][i]=dp[][i-]+s[k['-']][k[str2[i-]]];
}
for(i=;i<=n1;i++)
for(j=;j<=n2;j++)
dp[i][j]=max(dp[i-][j-]+s[k[str1[i-]]][k[str2[j-]]],dp[i][j-]+s[k['-']][k[str2[j-]]],dp[i-][j]+s[k[str1[i-]]][k['-']]);
t--;
cout<<dp[n1][n2]<<endl;
}
return ;
}
杭电20题 Human Gene Functions的更多相关文章
- 杭电1080 J - Human Gene Functions
题目大意: 两个字符串,可以再中间任何插入空格,然后让这两个串匹配,字符与字符之间的匹配有各自的分数,求最大分数 最长公共子序列模型. dp[i][j]表示当考虑吧串1的第i个字符和串2的第j个字符时 ...
- poj 1080 ——Human Gene Functions——————【最长公共子序列变型题】
Human Gene Functions Time Limit: 1000MS Memory Limit: 10000K Total Submissions: 17805 Accepted: ...
- poj 1080 Human Gene Functions(lcs,较难)
Human Gene Functions Time Limit: 1000MS Memory Limit: 10000K Total Submissions: 19573 Accepted: ...
- POJ 1080:Human Gene Functions LCS经典DP
Human Gene Functions Time Limit: 1000MS Memory Limit: 10000K Total Submissions: 18007 Accepted: ...
- 高手看了,感觉惨不忍睹——关于“【ACM】杭电ACM题一直WA求高手看看代码”
按 被中科大软件学院二年级研究生 HCOONa 骂为“误人子弟”之后(见:<中科大的那位,敢更不要脸点么?> ),继续“误人子弟”. 问题: 题目:(感谢 王爱学志 网友对题目给出的翻译) ...
- Human Gene Functions
Human Gene Functions Time Limit: 1000MS Memory Limit: 10000K Total Submissions: 18053 Accepted: 1004 ...
- Help Johnny-(类似杭电acm3568题)
Help Johnny(类似杭电3568题) Description Poor Johnny is so busy this term. His tutor threw lots of hard pr ...
- hdu1080 Human Gene Functions() 2016-05-24 14:43 65人阅读 评论(0) 收藏
Human Gene Functions Time Limit: 2000/1000 MS (Java/Others) Memory Limit: 65536/32768 K (Java/Oth ...
- 【POJ 1080】 Human Gene Functions
[POJ 1080] Human Gene Functions 相似于最长公共子序列的做法 dp[i][j]表示 str1[i]相应str2[j]时的最大得分 转移方程为 dp[i][j]=max(d ...
随机推荐
- 设计模式 -- 代理模式 (Proxy Pattern)
定义: 为其他对象提供一种代理以控制对这个对象的访问: 角色: 1,抽象主题类,(接口或者抽象类),抽象真实主题和代理的共有方法(如下Subject类): 2,具体实现的主题类,继承或者实现抽象主题类 ...
- [河南省ACM省赛-第三届] 房间安排 (nyoj 168)
题目链接:http://acm.nyist.net/JudgeOnline/problem.php?pid=168 分析:找到一天中需要最多的房间即可 #include<iostream> ...
- 移动HTML 5前端性能优化指南(转载)
前端工程师的菜!最近移动Html 5越来越火,想有一个体验流畅的Html 5 应用,这篇优化指南就别放过咯.腾讯的同学将关键的注意点与优化方法都总结出来,全文高能干货,非常值得深度学习 >> ...
- Linux 服务器系统监控脚本 Shell【转】
转自: Linux 服务器系统监控脚本 Shell - 今日头条(www.toutiao.com)http://www.toutiao.com/i6373134402163048961/ 本程序在Ce ...
- Linux 查公网出口IP
wget http://members.3322.org/dyndns/getipcat getip
- PHP mktime函数获取今天的开始和结束时间戳
php 获取今日.昨日.上周.本月的起始时间戳和结束时间戳的方法,主要使用到了 php 的时间函数 mktime(). mktime函数用法如下:mktime(hour,minute,second,m ...
- 初识Spark(Spark系列)
1.Spark Spark是继Hadoop之后,另外一种开源的高效大数据处理引擎,目前已提交为apach顶级项目. 效率: 据官方网站介绍,Spark是Hadoop运行效率的10-100倍(随内存计算 ...
- 如何判断Socket已经关闭
引子 前段时间我们的服务由于一台交换机网络出现故障,导致数据库连接不上,但是在数据库的连接超时参数设置不合理,connect timeout设置的过长,导致接口耗时增加.DB连接超时后线程未正常结束, ...
- Android原生Cling演化的覆盖层
package com.example.demotest; import android.content.Context; import android.graphics.Paint; import ...
- 关于设置CFileDialog的默认路径
CFileDialog d_File(FRUE, NULL,NULL,NULL,szFilter,FromHandle(m_hWnd)); // 如果写了下面这句那么每次打开都是这个设置的默认路径 ...
