dp-(LCS 基因匹配)
| Time Limit: 1000MS | Memory Limit: 10000K | |
| Total Submissions: 19885 | Accepted: 11100 |
Description
A human gene can be identified through a series of
time-consuming biological experiments, often with the help of computer
programs. Once a sequence of a gene is obtained, the next job is to
determine its function.
One of the methods for biologists to use in determining the function
of a new gene sequence that they have just identified is to search a
database with the new gene as a query. The database to be searched
stores many gene sequences and their functions – many researchers have
been submitting their genes and functions to the database and the
database is freely accessible through the Internet.
A database search will return a list of gene sequences from the database that are similar to the query gene.
Biologists assume that sequence similarity often implies
functional similarity. So, the function of the new gene might be
one of the functions that the genes from the list have. To exactly
determine which one is the right one another series of biological
experiments will be needed.
Your job is to make a program that compares two genes and determines
their similarity as explained below. Your program may be used as a part
of the database search if you can provide an efficient one.
Given two genes AGTGATG and GTTAG, how similar are they? One of the methods to measure the similarity
of two genes is called alignment. In an alignment, spaces are inserted, if necessary, in appropriate positions of
the genes to make them equally long and score the resulting genes according to a scoring matrix.
For example, one space is inserted into AGTGATG to result in
AGTGAT-G, and three spaces are inserted into GTTAG to result in
–GT--TAG. A space is denoted by a minus sign (-). The two genes
are now of equal
length. These two strings are aligned:
AGTGAT-G
-GT--TAG
In this alignment, there are four matches, namely, G in the second
position, T in the third, T in the sixth, and G in the eighth. Each
pair of aligned characters is assigned a score according to the
following scoring matrix.
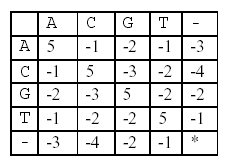
denotes that a space-space match is not allowed. The score
of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9.
Of course, many other alignments are possible. One is shown below (a
different number of spaces are inserted into different positions):
AGTGATG
-GTTA-G
This alignment gives a score of (-3)+5+5+(-2)+5+(-1) +5=14. So,
this one is better than the previous one. As a matter of fact, this one
is optimal since no other alignment can have a higher score. So, it is
said that the
similarity of the two genes is 14.
Input
input consists of T test cases. The number of test cases ) (T is
given in the first line of the input file. Each test case consists of
two lines: each line contains an integer, the length of a gene, followed
by a gene sequence. The length of each gene sequence is at least one
and does not exceed 100.
Output
Sample Input
2
7 AGTGATG
5 GTTAG
7 AGCTATT
9 AGCTTTAAA
Sample Output
14
21 题目大意 : 给出一个基因匹配表格 , 里面有一些数值 。
设 dp[i][j] 为 s1 取第 i 个字符, s2 取第 j 个字符的最大值,决定dp[i][j] 最优的情况有 三种, 类似于最长公共子序列的三种情况。
1 . s1 取第 i 个字母, s2 取 '-' , temp1 = dp[i-1][j] + score[a[i]]['-'];
2 . s2 取第 j 个字母, s1 取 '-' , temp2 = dp[i][j-1] + score['-'][b[j]];
3 . s1 取第 i 个字母 , s2 取第 j 个字母 , temp3 = dp[i-1][j-1] + score[a[i]][b[j]]; 则 dp[i][j] = max ( temp1, temp2, temp3 ); 还有初始化问题 : dp[0][0] = 0;
dp[i][0] = dp[i-1][0] + score[a[i]]['-'];
dp[0][j] = dp[0][j-1] + score['-'][b[j]]; 代码示例 :
/*
* Author: ry
* Created Time: 2017/9/3 8:00:06
* File Name: 1.cpp
*/
#include <iostream>
#include <cstdio>
#include <cstdlib>
#include <cstring>
#include <cmath>
#include <algorithm>
#include <string>
#include <vector>
#include <stack>
#include <queue>
#include <set>
#include <time.h>
using namespace std;
const int mm = 1e6+5;
#define ll long long ll t_cnt;
void t_st(){t_cnt=clock();}
void t_ot(){printf("you spent : %lldms\n", clock()-t_cnt);}
//开始t_st();
//结束t_ot(); int dp[150][150];
int score['T'+1]['T'+1]; void intial () {
score['A']['A'] = 5;
score['C']['C'] = 5;
score['G']['G'] = 5;
score['T']['T'] = 5;
score['A']['C'] = score['C']['A'] = -1;
score['A']['G'] = score['G']['A'] = -2;
score['A']['T'] = score['T']['A'] = -1;
score['A']['-'] = score['-']['A'] = -3;
score['C']['G'] = score['G']['C'] = -3;
score['C']['T'] = score['T']['C'] = -2;
score['C']['-'] = score['-']['C'] = -4;
score['G']['T'] = score['T']['G'] = -2;
score['G']['-'] = score['-']['G'] = -2;
score['T']['-'] = score['-']['T'] = -1;
} int MAX (int x, int y, int z) {
int k = (x>y?x:y);
return z>k?z:k;
} int main() {
int t;
int len1, len2;
char a[105], b[105]; intial();
cin >> t;
getchar();
while ( t-- ){
memset (dp, 0, sizeof(dp));
scanf("%d %s", &len1, a);
scanf("%d %s", &len2, b); for (int i = len1; i > 0; i--)
a[i] = a[i-1];
for (int i = len2; i > 0; i--)
b[i] = b[i-1]; dp[0][0] = 0;
for (int i = 1; i <= len1; i++)
dp[i][0] = dp[i-1][0] + score[a[i]]['-'];
for (int j = 1; j <= len2; j++)
dp[0][j] = dp[0][j-1] + score['-'][b[j]]; for (int i = 1; i <= len1; i++)
for (int j = 1; j <= len2; j++){
int temp1 = dp[i-1][j] + score[a[i]]['-'];
int temp2 = dp[i][j-1] + score['-'][b[j]];
int temp3 = dp[i-1][j-1] + score[a[i]][b[j]];
dp[i][j] = MAX (temp1, temp2, temp3);
} printf ("%d\n", dp[len1][len2]);
} return 0;
}
dp-(LCS 基因匹配)的更多相关文章
- 【线型DP】【LCS】洛谷P4303 [AHOI2006]基因匹配
P4303 [AHOI2006]基因匹配 标签(空格分隔): 考试题 nt题 LCS优化 [题目] 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球 ...
- BZOJ 1264: [AHOI2006]基因匹配Match( LCS )
序列最大长度2w * 5 = 10w, O(n²)的LCS会T.. LCS 只有当a[i] == b[j]时, 才能更新答案, 我们可以记录n个数在第一个序列中出现的5个位置, 然后从左往右扫第二个序 ...
- bzoj 1264 [AHOI2006]基因匹配Match(DP+树状数组)
1264: [AHOI2006]基因匹配Match Time Limit: 10 Sec Memory Limit: 162 MBSubmit: 793 Solved: 503[Submit][S ...
- bzoj1264 [AHOI2006]基因匹配Match 树状数组+lcs
1264: [AHOI2006]基因匹配Match Time Limit: 10 Sec Memory Limit: 162 MBSubmit: 1255 Solved: 835[Submit][ ...
- BZOJ 1264: [AHOI2006]基因匹配Match 树状数组+DP
1264: [AHOI2006]基因匹配Match Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球 ...
- 【BZOJ1264】[AHOI2006]基因匹配Match DP+树状数组
[BZOJ1264][AHOI2006]基因匹配Match Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而 ...
- 基因匹配(bzoj 1264)
Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球上只有4种),而更奇怪的是,组成DNA序列的每一种碱 ...
- BZOJ1264: [AHOI2006]基因匹配Match
1264: [AHOI2006]基因匹配Match Time Limit: 10 Sec Memory Limit: 162 MBSubmit: 541 Solved: 347[Submit][S ...
- [AHOI2006]基因匹配
题目描述 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球上只有4种),而更奇怪的是,组成DNA序列的每一种碱基在该序列中正好出现5次!这样如果一个 ...
- bzoj 1264: [AHOI2006]基因匹配Match
1264: [AHOI2006]基因匹配Match Description 基因匹配(match) 卡卡昨天晚上做梦梦见他和可可来到了另外一个星球,这个星球上生物的DNA序列由无数种碱基排列而成(地球 ...
随机推荐
- webpack学习(一)项目中安装webpack
如何在项目中安装webpack,webpack-cli? 前提:电脑安装了 node和npm包管理工具 1 创建项目文件夹或者在已有的项目中打开终端 输入相关命令: npm init 因为已经安装好 ...
- vue——父子传值
转载地址:https://blog.csdn.net/xr510002594/article/details/83304141
- requires php ~7.1 -> your PHP version (7.0.18) does not satisfy that requirement
一个大兄弟本地用了 PHP 7.1 进行开发,而我本地是 PHP 7.0, 于是悲剧发生了. composer install 之后报错 Loading composer repositories w ...
- 微信小程序之在线试题(1)
最近在做一套公司的市场化培训项目,涉及到手机端在线答题的设计,首先摒弃app的模式,那就只剩下微信公众号和小程序,而公众号是可以关联小程序,所以我们只需要做好一套小程序. 因为篇幅问题,下面只讲解在线 ...
- 【t066】致命的珠宝
Time Limit: 1 second Memory Limit: 128 MB [问题描述] 门上有着N个宝珠,每个宝珠都有一个数字.Mini询问老者后,得知要想打开这扇门,就得找出两颗珠宝,使这 ...
- Web中的通配符
/**的意思是所有文件夹及里面的子文件夹 /*是所有文件夹,不含子文件夹 /是web项目的根目录 http://www.coderanch.com/t/364782/Servlets/java ...
- How to output the target message in dotnet build command line
How can I output my target message when I using dotnet build in command line. I use command line to ...
- X-Admin&ABP框架开发-代码生成器
在日常开发中,有时会遇到一些相似的代码,甚至是只要CV一次,改几个名称,就可以实现功能了,而且总归起来,都可以由一些公用的页面更改而来,因此,结合我日常开发中使用到的页面,封装一个适合自己的代码生成器 ...
- nginx负载均衡的相关配置
一台nginx的负载均衡服务器(172.25.254.131) 两台安装httpd作为web端 一.准备工作 1.1 安装nginx yum -y install gcc openssl-devel ...
- 面试官刁难:Java字符串可以引用传递吗?
老读者都知道了,六年前,我从苏州回到洛阳,抱着一幅"海归"的心态,投了不少简历,也"约谈"了不少面试官,但仅有两三个令我感到满意.其中有一位叫老马,至今还活在我 ...
