HDU1560 DNA sequence —— IDA*算法
题目链接:http://acm.hdu.edu.cn/showproblem.php?pid=1560
DNA sequence
Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others)
Total Submission(s): 2999 Accepted Submission(s): 1462
sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of the given sequence is the subsequence
of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
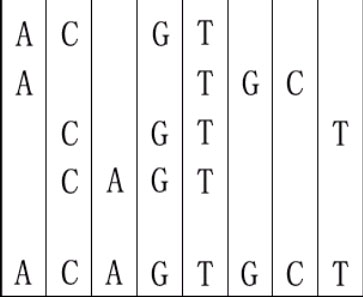
sequence is between 1 and 5.
4
ACGT
ATGC
CGTT
CAGT
题解:
一开始以为是直接用回溯的方法,结果TLE。看了题解是用IDA*(迭代加深搜),其实自己不太了解迭代加深搜为什么比较快,而且什么时候用合适?下面是自己对迭代加深搜的一些浅薄的了解:
1.首先迭代加深搜适合用在:求最少步数(带有BFS的特点)并且不太容易估计搜索深度的问题上,同时兼有了BFS求最少步数和DFS易写、无需多开数组的特点。
2.相对于赤裸裸的回溯,迭代加深搜由于限制了搜索深度,所以也能适当地剪枝。
3.我编不下去了……
代码一:
#include <iostream>
#include <cstdio>
#include <cstring>
#include <cmath>
#include <algorithm>
#include <vector>
#include <queue>
#include <stack>
#include <map>
#include <string>
#include <set>
#define ms(a,b) memset((a),(b),sizeof((a)))
using namespace std;
typedef long long LL;
const int INF = 2e9;
const LL LNF = 9e18;
const int MOD = 1e9+;
const int MAXN = +; int n;
char dna[MAXN][MAXN];
int len[MAXN], pos[MAXN];
char s[] = {'A', 'G', 'C', 'T'}; bool dfs(int k, int limit) //k为放了几个, k+1才为当前要放的
{
int maxx = , cnt = ; //maxx为最长剩余的dna片段, cnt为剩余的片段之和(核苷酸链?好怀念啊)
for(int i = ; i<n; i++)
{
cnt += len[i]-pos[i];
maxx = max(maxx, len[i]-pos[i]);
}
if(cnt==) return true; //如果片段都放完,则已得到答案
if(cnt<=limit-k) return true; //剪枝:片段之和小于等于剩余能放数量,肯定能够得到答案
if(maxx>limit-k) return false; //剪枝:最小的估计值都大于剩余能放数量,肯定不能得到答案 int tmp[MAXN];
for(int i = ; i<; i++)
{
memcpy(tmp, pos, sizeof(tmp));
bool flag = false;
for(int j = ; j<n; j++)
if(dna[j][pos[j]]==s[i])
pos[j]++, flag = true; //k+1<=limit:在限制范围内
if(k+<=limit && flag && dfs(k+, limit) )
return true;
memcpy(pos, tmp, sizeof(pos));
}
return false;
} int main()
{
int T;
scanf("%d",&T);
while(T--)
{
scanf("%d",&n);
int limit = ;
for(int i = ; i<n; i++)
{
scanf("%s",dna[i]);
len[i] = strlen(dna[i]);
limit = max(limit, len[i]);
} ms(pos, );
while(!dfs(, limit))
limit++;
printf("%d\n", limit);
}
}
代码二:
#include <iostream>
#include <cstdio>
#include <cstring>
#include <cmath>
#include <algorithm>
#include <vector>
#include <queue>
#include <stack>
#include <map>
#include <string>
#include <set>
#define ms(a,b) memset((a),(b),sizeof((a)))
using namespace std;
typedef long long LL;
const int INF = 2e9;
const LL LNF = 9e18;
const int MOD = 1e9+;
const int MAXN = +; int n;
char dna[MAXN][MAXN];
int len[MAXN], pos[MAXN];
char s[] = {'A', 'G', 'C', 'T'}; bool dfs(int k, int limit) //k为放了几个, k+1才为当前要放的
{
if(k>limit) return false; int maxx = , cnt = ; //maxx为最长剩余的dna片段, cnt为剩余的片段之和(核苷酸链?好怀念啊)
for(int i = ; i<n; i++)
{
cnt += len[i]-pos[i];
maxx = max(maxx, len[i]-pos[i]);
}
if(cnt==) return true; //如果片段都放完,则已得到答案
if(cnt<=limit-k) return true; //剪枝:片段之和小于等于剩余能放数量,肯定能够得到答案
if(maxx>limit-k) return false; //剪枝:最小的估计值都大于剩余能放数量,肯定不能得到答案 int tmp[MAXN];
for(int i = ; i<; i++)
{
memcpy(tmp, pos, sizeof(tmp));
bool flag = false;
for(int j = ; j<n; j++)
if(dna[j][pos[j]]==s[i])
pos[j]++, flag = true; if(flag && dfs(k+, limit) )
return true;
memcpy(pos, tmp, sizeof(pos));
}
return false;
} int main()
{
int T;
scanf("%d",&T);
while(T--)
{
scanf("%d",&n);
int limit = ;
for(int i = ; i<n; i++)
{
scanf("%s",dna[i]);
len[i] = strlen(dna[i]);
limit = max(limit, len[i]);
} ms(pos, );
while(!dfs(, limit))
limit++;
printf("%d\n", limit);
}
}
HDU1560 DNA sequence —— IDA*算法的更多相关文章
- HDU1560 DNA sequence IDA* + 强力剪枝 [kuangbin带你飞]专题二
题意:给定一些DNA序列,求一个最短序列能够包含所有序列. 思路:记录第i个序列已经被匹配的长度p[i],以及第i序列的原始长度len[i].则有两个剪枝: 剪枝1:直接取最长待匹配长度.1900ms ...
- HDU1560 DNA sequence(IDA*)题解
DNA sequence Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others) To ...
- Hdu1560 DNA sequence(IDA*) 2017-01-20 18:53 50人阅读 评论(0) 收藏
DNA sequence Time Limit : 15000/5000ms (Java/Other) Memory Limit : 32768/32768K (Java/Other) Total ...
- HDU1560 DNA sequence
题目: The twenty-first century is a biology-technology developing century. We know that a gene is made ...
- HDU 1560 DNA sequence (IDA* 迭代加深 搜索)
题目地址:http://acm.hdu.edu.cn/showproblem.php?pid=1560 BFS题解:http://www.cnblogs.com/crazyapple/p/321810 ...
- HDU 1560 DNA sequence(IDA*)
题目链接:http://acm.hdu.edu.cn/showproblem.php?pid=1560 题目大意:给出n个字符串,让你找一个字符串使得这n个字符串都是它的子串,求最小长度. 解题思路: ...
- DNA sequence HDU - 1560(IDA*,迭代加深搜索)
题目大意:有n个DNA序列,构造一个新的序列,使得这n个DNA序列都是它的子序列,然后输出最小长度. 题解:第一次接触IDA*算法,感觉~~好暴力!!思路:维护一个数组pos[i],表示第i个串该匹配 ...
- 【学时总结】 ◆学时·II◆ IDA*算法
[学时·II] IDA*算法 ■基本策略■ 如果状态数量太多了,优先队列也难以承受:不妨再回头看DFS-- A*算法是BFS的升级,那么IDA*算法是对A*算法的再优化,同时也是对迭代加深搜索(IDF ...
- DNA sequence(映射+BFS)
Problem Description The twenty-first century is a biology-technology developing century. We know tha ...
随机推荐
- JavaScript 的时间消耗--摘抄
JavaScript 的时间消耗 2017-12-24 dwqs 前端那些事儿 随着我们的网站越来越依赖 JavaScript, 我们有时会(无意)用一些不易追踪的方式来传输一些(耗时的)东西. 在这 ...
- CSV文件导出2
public void exportCSVFile( HttpServletResponse response, ResultSet rs,String fileName,String headers ...
- grequests----golang的requests库
github.com/levigross/grequests: A Go "clone" of the great and famous Requests library 特点: ...
- poj2513字典树+欧拉图判断+并查集断连通
题意:俩头带有颜色的木棒,要求按颜色同的首尾相连,可能否? 思路:棒子本身是一条边,以俩端为顶点(同颜色共点),即求是否有无向图欧拉路(每条棒子只有一根, 边只能用一次,用一次边即选一次棒子). 先判 ...
- LeetCode OJ--Valid Palindrome
http://oj.leetcode.com/problems/valid-palindrome/ 判断是否为回文串 bool isPalindrome(string s) { ,j = s.leng ...
- grafana 安装 和 Nginx 、EL 联调
https://blog.csdn.net/u010735147/article/details/80943593
- 安装Django时解决的问题-mysql及访问(附pycharm激活)
1.做些软链接和virtualenv的基本使用: ln -s /data/linkdood/im/vrv/python36/bin/python3.6 /usr/bin/python3 ln -s / ...
- cut printf awk sed grep笔记
名称 作用 参数 实例 cut 截取某列,可指定分隔 -f 列号 -d 分隔符 cut -d ":" -f 1, 3 /etc/passwd 截取第一列和第三列 printf pr ...
- CentOS6.5升级手动安装GCC4.8.2 与 CentOS 6.4 编译安装 gcc 4.8.1
http://blog.163.com/zhu329599788@126/blog/static/6669335020161179259975 http://www.cnblogs.com/codem ...
- 加密算法和MD5等散列算法的区别(转)
本文转自http://www.cnblogs.com/eternalwt/archive/2013/03/21/2973807.html 感谢作者 1.在软件开发的用户注册功能中常出现MD5加密这个概 ...
