uva1368 DNA Consensus String
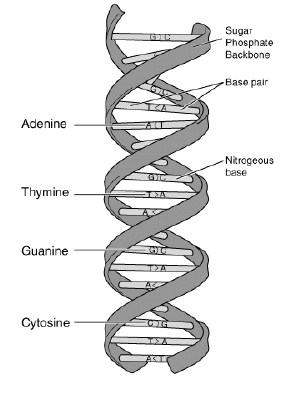 <tex2html_verbatim_mark> Figure 1.
<tex2html_verbatim_mark> Figure 1.DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It consists of four different nucleotides, namely Adenine, Thymine, Guanine, and Cytosine as shown in Figure 1. If we represent a nucleotide by its initial character, a DNA strand can be regarded as a long string (sequence of characters) consisting of the four characters A, T, G, and C. For example, assume we are given some part of a DNA strand which is composed of the following sequence of nucleotides:
``Thymine-Adenine-Adenine-Cytosine-Thymine-Guanine-Cytosine-Cytosine-Guanine-Adenine-Thymine"
Then we can represent the above DNA strand with the string ``TAACTGCCGAT." The biologist Prof. Ahn found that a gene X commonly exists in the DNA strands of five different kinds of animals, namely dogs, cats, horses, cows, and monkeys. He also discovered that the DNA sequences of the gene X from each animal were very alike. See Figure 2.
| DNA sequence of gene X | |
| Cat: | GCATATGGCTGTGCA |
| Dog: | GCAAATGGCTGTGCA |
| Horse: | GCTAATGGGTGTCCA |
| Cow: | GCAAATGGCTGTGCA |
| Monkey: | GCAAATCGGTGAGCA |
Prof. Ahn thought that humans might also have the gene X and decided to search for the DNA sequence of X in human DNA. However, before searching, he should define a representative DNA sequence of gene X because its sequences are not exactly the same in the DNA of the five animals. He decided to use the Hamming distance to define the representative sequence. The Hamming distance is the number of different characters at each position from two strings of equal length. For example, assume we are given the two strings ``AGCAT" and ``GGAAT." The Hamming distance of these two strings is 2 because the 1st and the 3rd characters of the two strings are different. Using the Hamming distance, we can define a representative string for a set of multiple strings of equal length. Given a set of strings S = s1,...,sm<tex2html_verbatim_mark> of length n<tex2html_verbatim_mark> , the consensus error between a string y<tex2html_verbatim_mark> of length n<tex2html_verbatim_mark> and the set S<tex2html_verbatim_mark> is the sum of the Hamming distances between y<tex2html_verbatim_mark> and eachsi<tex2html_verbatim_mark> in S<tex2html_verbatim_mark> . If the consensus error between y<tex2html_verbatim_mark> and S<tex2html_verbatim_mark> is the minimum among all possible strings y<tex2html_verbatim_mark> of length n<tex2html_verbatim_mark> , y<tex2html_verbatim_mark> is called a consensus string ofS<tex2html_verbatim_mark> . For example, given the three strings `` AGCAT" `` AGACT" and `` GGAAT" the consensus string of the given strings is `` AGAAT" because the sum of the Hamming distances between `` AGAAT" and the three strings is 3 which is minimal. (In this case, the consensus string is unique, but in general, there can be more than one consensus string.) We use the consensus string as a representative of the DNA sequence. For the example of Figure 2 above, a consensus string of gene X is `` GCAAATGGCTGTGCA" and the consensus error is 7.
Input
Your program is to read from standard input. The input consists of T<tex2html_verbatim_mark> test cases. The number of test cases T<tex2html_verbatim_mark> is given in the first line of the input. Each test case starts with a line containing two integers m<tex2html_verbatim_mark> and n<tex2html_verbatim_mark> which are separated by a single space. The integer m<tex2html_verbatim_mark>(4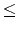 m
m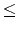 50)<tex2html_verbatim_mark>represents the number of DNA sequences and n<tex2html_verbatim_mark>(4
50)<tex2html_verbatim_mark>represents the number of DNA sequences and n<tex2html_verbatim_mark>(4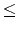 n
n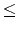 1000)<tex2html_verbatim_mark> represents the length of the DNA sequences, respectively. In each of the next m<tex2html_verbatim_mark> lines, each DNA sequence is given.
1000)<tex2html_verbatim_mark> represents the length of the DNA sequences, respectively. In each of the next m<tex2html_verbatim_mark> lines, each DNA sequence is given.
Output
Your program is to write to standard output. Print the consensus string in the first line of each case and the consensus error in the second line of each case. If there exists more than one consensus string, print the lexicographically smallest consensus string. The following shows sample input and output for three test cases.
Sample Input
3
5 8
TATGATAC
TAAGCTAC
AAAGATCC
TGAGATAC
TAAGATGT
4 10
ACGTACGTAC
CCGTACGTAG
GCGTACGTAT
TCGTACGTAA
6 10
ATGTTACCAT
AAGTTACGAT
AACAAAGCAA
AAGTTACCTT
AAGTTACCAA
TACTTACCAA
Sample Output
TAAGATAC
7
ACGTACGTAA
6
AAGTTACCAA
12
这个问题比较简单,注意字典序就好了。
附上AC代码:
#include<iostream>
#include<algorithm> using namespace std;
char str[][];
char ch[];
int t[];
int n;
int a,b;
int Sum;
int Flag;
int main()
{
cin >> n;
while(n--)
{
Sum=;
cin >> a >> b;
for(int i=;i<a;i++)
cin >> str[i];
for(int i=;i<b;i++)
{
Flag=;
t[]=t[]=t[]=t[]=;
for(int j=;j<a;j++)
{
if(str[j][i]=='A') t[]++;
if(str[j][i]=='C') t[]++;
if(str[j][i]=='G') t[]++;
if(str[j][i]=='T') t[]++;
}
for(int i=;i<;i++)
if(t[i]>t[Flag]) Flag=i;
if(Flag==) cout << 'A';
if(Flag==) cout << 'C';
if(Flag==) cout << 'G';
if(Flag==) cout << 'T';
Sum+=t[Flag];
}
cout << endl;
cout << a*b-Sum << endl;
}
return ;
}
uva1368 DNA Consensus String的更多相关文章
- UVA-1368 DNA Consensus String(思路)
题目: 链接 题意: 题目虽然比较长,但读完之后题目的思路还是比较容易想出来的. 给出m个长度为n的字符串(只包含‘A’.‘T’.‘G’.‘C’),我们的任务是得出一个字符串,要求这个字符串与给出的m ...
- 贪心水题。UVA 11636 Hello World,LA 3602 DNA Consensus String,UVA 10970 Big Chocolate,UVA 10340 All in All,UVA 11039 Building Designing
UVA 11636 Hello World 二的幂答案就是二进制长度减1,不是二的幂答案就是是二进制长度. #include<cstdio> int main() { ; ){ ; ) r ...
- DNA Consensus String
题目(中英对照): DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It co ...
- 紫书第三章训练1 E - DNA Consensus String
DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It consists of ...
- UVa1368/ZOJ3132 DNA Consensus String
#include <stdio.h>#include <string.h> int main(){ int a[4][1000]; // A/C/G/T在每列中出现的次数 ...
- DNA序列 (DNA Consensus String,ACM/ICPC Seoul 2006,UVa1368
题目描述:算法竞赛入门经典习题3-7 题目思路:每列出现最多的距离即最短 #include <stdio.h> #include <string.h> int main(int ...
- uva 1368 DNA Consensus String
这道题挺简单的,刚开始理解错误,以为是从已有的字符串里面求最短的距离,后面才发现是求一个到所有字符串最小距离的字符串,因为这样的字符串可能有多个,所以最后取最小字典序的字符串. 我的思路就是求每一列每 ...
- uvalive 3602 DNA Consensus String
https://vjudge.net/problem/UVALive-3602 题意: 给定m个长度均为n的DNA序列,求一个DNA序列,使得它到所有的DNA序列的汉明距离最短,若有多个解则输出字典序 ...
- 【每日一题】UVA - 1368 DNA Consensus String 字符串+贪心+阅读题
https://cn.vjudge.net/problem/UVA-1368 二维的hamming距离算法: For binary strings a and b the Hamming distan ...
随机推荐
- 关于SRAM,DRAM,SDRAM,以及NORFLASH,NANDFLASH
韦东山的视频里面说S3C2440有4KB的内存,这个其实是不正确的,这4KB的RAM严格说不应该叫内存,严格来说芯片外面的64MB的SDRAM才能叫做内存,里面的那4KB只是当nandflash启动的 ...
- python编程之处理GB级的大型文件
一般我们采取分块处理,一次处理固定大小的块. def read_in_chunks(file_obj,chunk_size): """Lazy function (gen ...
- using 1.7 requires using android build tools version 19 or later
这意思大概是adt用了1.7,abt(android build tools)就要用19或更高,可是abt在哪设置呢,原来是在sdk manager中 之前我已安装的最高的abt是17,然后~~~,F ...
- VMware Workstation 精致汉化系列 使用方法
http://kuai.xunlei.com/d/QqGABAKChQBwMzxR983 迅雷快传 XP系统之家-温馨提示: VMware Workstation 精致汉化系列 使用方法:1.安装 ...
- 批量更新sql |批量update sql
图所示现需要批量更新table2表内字段Pwd更新userName对IP地址username与Ip对应关系table1所示 update table2 set pwd=table1.ip from t ...
- Java常见内存溢出异常分析(OutOfMemoryError)
原文转载自:http://my.oschina.net/sunchp/blog/369412 1.背景知识 1).JVM体系结构 2).JVM运行时数据区 JVM内存结构的相关可以参考: http:/ ...
- swift 点击button改变其内填充图片,达到选中的效果
先看下效果: 点击后: 实现:在页面拖一个button,然后在所在页面声明其变量和一个点击事件 声明: @IBOutlet weak var BtnZiDong: UIButton! 点击事件函数: ...
- APP制作过程
直播App开发的过程 第一步:分解直播App的功能,我们以X客为例 视频直播功能,这是一款直播App最主要的功能,要能支持视频直播RTMP推流,使画面传输流畅.清晰(美颜后的清晰,你懂的聊天功能,用户 ...
- [RxJS] Reactive Programming - Using cached network data with RxJS -- withLatestFrom()
So now we want to replace one user when we click the 'x' button. To do that, we want: 1. Get the cac ...
- 一个不错的PPT,扁平化设计,开放资源,要的进来
开了那么多的博客,没做啥资源贡献,今天共享一个不错的PPT模板.例如以下图所看到的,须要的话留下邮箱 watermark/2/text/aHR0cDovL2Jsb2cuY3Nkbi5uZXQvbGFp ...
