吴裕雄--天生自然 R语言开发学习:回归(续一)















#------------------------------------------------------------#
# R in Action (2nd ed): Chapter 8 #
# Regression #
# requires packages car, gvlma, MASS, leaps to be installed #
# install.packages(c("car", "gvlma", "MASS", "leaps")) #
#------------------------------------------------------------# par(ask=TRUE)
opar <- par(no.readonly=TRUE) # Listing 8.1 - Simple linear regression
fit <- lm(weight ~ height, data=women)
summary(fit)
women$weight
fitted(fit)
residuals(fit)
plot(women$height,women$weight,
main="Women Age 30-39",
xlab="Height (in inches)",
ylab="Weight (in pounds)")
# add the line of best fit
abline(fit) # Listing 8.2 - Polynomial regression
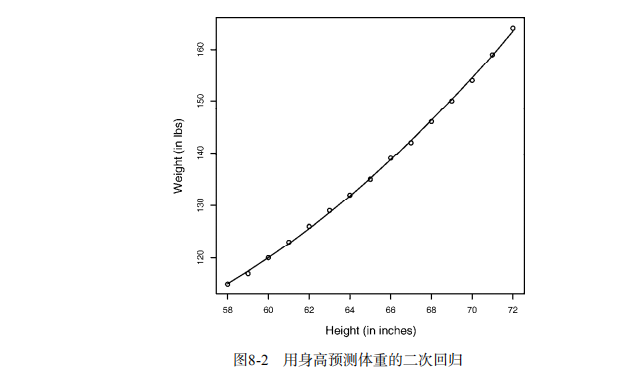
fit2 <- lm(weight ~ height + I(height^2), data=women)
summary(fit2)
plot(women$height,women$weight,
main="Women Age 30-39",
xlab="Height (in inches)",
ylab="Weight (in lbs)")
lines(women$height,fitted(fit2)) # Enhanced scatterplot for women data
library(car)
library(car)
scatterplot(weight ~ height, data=women,
spread=FALSE, smoother.args=list(lty=2), pch=19,
main="Women Age 30-39",
xlab="Height (inches)",
ylab="Weight (lbs.)") # Listing 8.3 - Examining bivariate relationships
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
cor(states)
library(car)
scatterplotMatrix(states, spread=FALSE, smoother.args=list(lty=2),
main="Scatter Plot Matrix") # Listing 8.4 - Multiple linear regression
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
fit <- lm(Murder ~ Population + Illiteracy + Income + Frost, data=states)
summary(fit) # Listing 8.5 - Mutiple linear regression with a significant interaction term
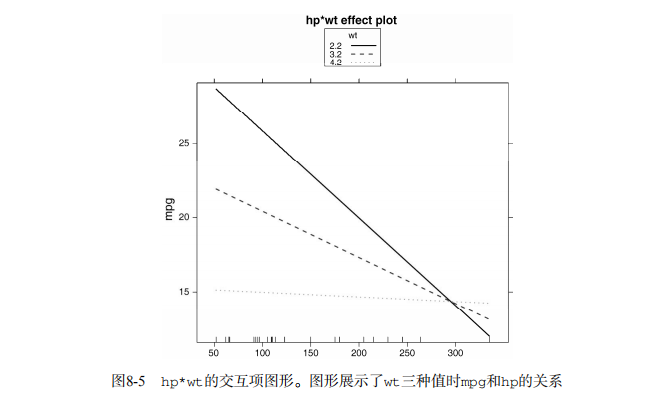
fit <- lm(mpg ~ hp + wt + hp:wt, data=mtcars)
summary(fit) library(effects)
plot(effect("hp:wt", fit,, list(wt=c(2.2, 3.2, 4.2))), multiline=TRUE) # simple regression diagnostics
fit <- lm(weight ~ height, data=women)
par(mfrow=c(2,2))
plot(fit)
newfit <- lm(weight ~ height + I(height^2), data=women)
par(opar)
par(mfrow=c(2,2))
plot(newfit)
par(opar) # basic regression diagnostics for states data
opar <- par(no.readonly=TRUE)
fit <- lm(weight ~ height, data=women)
par(mfrow=c(2,2))
plot(fit)
par(opar) fit2 <- lm(weight ~ height + I(height^2), data=women)
opar <- par(no.readonly=TRUE)
par(mfrow=c(2,2))
plot(fit2)
par(opar) # Assessing normality
library(car)
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
fit <- lm(Murder ~ Population + Illiteracy + Income + Frost, data=states)
qqPlot(fit, labels=row.names(states), id.method="identify",
simulate=TRUE, main="Q-Q Plot") # Listing 8.6 - Function for plotting studentized residuals
residplot <- function(fit, nbreaks=10) {
z <- rstudent(fit)
hist(z, breaks=nbreaks, freq=FALSE,
xlab="Studentized Residual",
main="Distribution of Errors")
rug(jitter(z), col="brown")
curve(dnorm(x, mean=mean(z), sd=sd(z)),
add=TRUE, col="blue", lwd=2)
lines(density(z)$x, density(z)$y,
col="red", lwd=2, lty=2)
legend("topright",
legend = c( "Normal Curve", "Kernel Density Curve"),
lty=1:2, col=c("blue","red"), cex=.7)
} residplot(fit) # Assessing linearity
library(car)
crPlots(fit) # Listing 8.7 - Assessing homoscedasticity
library(car)
ncvTest(fit)
spreadLevelPlot(fit) # Listing 8.8 - Global test of linear model assumptions
library(gvlma)
gvmodel <- gvlma(fit)
summary(gvmodel) # Listing 8.9 - Evaluating multi-collinearity
library(car)
vif(fit)
sqrt(vif(fit)) > 2 # problem? # Assessing outliers
library(car)
outlierTest(fit) # Identifying high leverage points
hat.plot <- function(fit) {
p <- length(coefficients(fit))
n <- length(fitted(fit))
plot(hatvalues(fit), main="Index Plot of Hat Values")
abline(h=c(2,3)*p/n, col="red", lty=2)
identify(1:n, hatvalues(fit), names(hatvalues(fit)))
}
hat.plot(fit) # Identifying influential observations # Cooks Distance D
# identify D values > 4/(n-k-1)
cutoff <- 4/(nrow(states)-length(fit$coefficients)-2)
plot(fit, which=4, cook.levels=cutoff)
abline(h=cutoff, lty=2, col="red") # Added variable plots
# add id.method="identify" to interactively identify points
library(car)
avPlots(fit, ask=FALSE, id.method="identify") # Influence Plot
library(car)
influencePlot(fit, id.method="identify", main="Influence Plot",
sub="Circle size is proportial to Cook's Distance" ) # Listing 8.10 - Box-Cox Transformation to normality
library(car)
summary(powerTransform(states$Murder)) # Box-Tidwell Transformations to linearity
library(car)
boxTidwell(Murder~Population+Illiteracy,data=states) # Listing 8.11 - Comparing nested models using the anova function
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
fit1 <- lm(Murder ~ Population + Illiteracy + Income + Frost,
data=states)
fit2 <- lm(Murder ~ Population + Illiteracy, data=states)
anova(fit2, fit1) # Listing 8.12 - Comparing models with the AIC
fit1 <- lm(Murder ~ Population + Illiteracy + Income + Frost,
data=states)
fit2 <- lm(Murder ~ Population + Illiteracy, data=states)
AIC(fit1,fit2) # Listing 8.13 - Backward stepwise selection
library(MASS)
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
fit <- lm(Murder ~ Population + Illiteracy + Income + Frost,
data=states)
stepAIC(fit, direction="backward") # Listing 8.14 - All subsets regression
library(leaps)
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
leaps <-regsubsets(Murder ~ Population + Illiteracy + Income +
Frost, data=states, nbest=4)
plot(leaps, scale="adjr2")
library(car)
subsets(leaps, statistic="cp",
main="Cp Plot for All Subsets Regression")
abline(1,1,lty=2,col="red") # Listing 8.15 - Function for k-fold cross-validated R-square
shrinkage <- function(fit,k=10){
require(bootstrap) # define functions
theta.fit <- function(x,y){lsfit(x,y)}
theta.predict <- function(fit,x){cbind(1,x)%*%fit$coef} # matrix of predictors
x <- fit$model[,2:ncol(fit$model)]
# vector of predicted values
y <- fit$model[,1] results <- crossval(x,y,theta.fit,theta.predict,ngroup=k)
r2 <- cor(y, fit$fitted.values)**2 # raw R2
r2cv <- cor(y,results$cv.fit)**2 # cross-validated R2
cat("Original R-square =", r2, "\n")
cat(k, "Fold Cross-Validated R-square =", r2cv, "\n")
cat("Change =", r2-r2cv, "\n")
} # using it
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
fit <- lm(Murder ~ Population + Income + Illiteracy + Frost, data=states)
shrinkage(fit)
fit2 <- lm(Murder~Population+Illiteracy,data=states)
shrinkage(fit2) # Calculating standardized regression coefficients
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
zstates <- as.data.frame(scale(states))
zfit <- lm(Murder~Population + Income + Illiteracy + Frost, data=zstates)
coef(zfit) # Listing 8.16 rlweights function for clculating relative importance of predictors
relweights <- function(fit,...){
R <- cor(fit$model)
nvar <- ncol(R)
rxx <- R[2:nvar, 2:nvar]
rxy <- R[2:nvar, 1]
svd <- eigen(rxx)
evec <- svd$vectors
ev <- svd$values
delta <- diag(sqrt(ev))
lambda <- evec %*% delta %*% t(evec)
lambdasq <- lambda ^ 2
beta <- solve(lambda) %*% rxy
rsquare <- colSums(beta ^ 2)
rawwgt <- lambdasq %*% beta ^ 2
import <- (rawwgt / rsquare) * 100
import <- as.data.frame(import)
row.names(import) <- names(fit$model[2:nvar])
names(import) <- "Weights"
import <- import[order(import),1, drop=FALSE]
dotchart(import$Weights, labels=row.names(import),
xlab="% of R-Square", pch=19,
main="Relative Importance of Predictor Variables",
sub=paste("Total R-Square=", round(rsquare, digits=3)),
...)
return(import)
} # Listing 8.17 - Applying the relweights function
states <- as.data.frame(state.x77[,c("Murder", "Population",
"Illiteracy", "Income", "Frost")])
fit <- lm(Murder ~ Population + Illiteracy + Income + Frost, data=states)
relweights(fit, col="blue")
吴裕雄--天生自然 R语言开发学习:回归(续一)的更多相关文章
- 吴裕雄--天生自然 R语言开发学习:R语言的安装与配置
下载R语言和开发工具RStudio安装包 先安装R
- 吴裕雄--天生自然 R语言开发学习:数据集和数据结构
数据集的概念 数据集通常是由数据构成的一个矩形数组,行表示观测,列表示变量.表2-1提供了一个假想的病例数据集. 不同的行业对于数据集的行和列叫法不同.统计学家称它们为观测(observation)和 ...
- 吴裕雄--天生自然 R语言开发学习:导入数据
2.3.6 导入 SPSS 数据 IBM SPSS数据集可以通过foreign包中的函数read.spss()导入到R中,也可以使用Hmisc 包中的spss.get()函数.函数spss.get() ...
- 吴裕雄--天生自然 R语言开发学习:使用键盘、带分隔符的文本文件输入数据
R可从键盘.文本文件.Microsoft Excel和Access.流行的统计软件.特殊格 式的文件.多种关系型数据库管理系统.专业数据库.网站和在线服务中导入数据. 使用键盘了.有两种常见的方式:用 ...
- 吴裕雄--天生自然 R语言开发学习:R语言的简单介绍和使用
假设我们正在研究生理发育问 题,并收集了10名婴儿在出生后一年内的月龄和体重数据(见表1-).我们感兴趣的是体重的分 布及体重和月龄的关系. 可以使用函数c()以向量的形式输入月龄和体重数据,此函 数 ...
- 吴裕雄--天生自然 R语言开发学习:基础知识
1.基础数据结构 1.1 向量 # 创建向量a a <- c(1,2,3) print(a) 1.2 矩阵 #创建矩阵 mymat <- matrix(c(1:10), nrow=2, n ...
- 吴裕雄--天生自然 R语言开发学习:图形初阶(续二)
# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Gettin ...
- 吴裕雄--天生自然 R语言开发学习:图形初阶(续一)
# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Gettin ...
- 吴裕雄--天生自然 R语言开发学习:图形初阶
# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Gettin ...
- 吴裕雄--天生自然 R语言开发学习:基本图形(续二)
#---------------------------------------------------------------# # R in Action (2nd ed): Chapter 6 ...
随机推荐
- Mariadb-10.2.25 多实例
Mariadb-10.2.25 多实例 定义目录 mkdir -p /mysql/{3306,3307,3308}/{bin,data,etc,log,pid,socket} 生成数据库文件 /app ...
- Win 10 Ctrl + Space 冲突
1. 说明 在IDE里面Ctrl + space 会与 Windows 输入法相互冲突,并且用Ctrl + Space 切换中英文也很不常用(常用直接shift切换). 2. 操作 控制面板——时钟. ...
- C++数组常用操作
1. 遍历数组 使用基于范围的for循环来遍历整个数组 用_countof()来得到数组中的元素个数 #include <iostream> #include <cstdio> ...
- 如何编写一个vue应用
1.vue应用的组成 1.1 vuex Vuex 是一个专为 Vue.js 应用程序开发的状态管理模式.它采用集中式存储管理应用的所有组件的状态,并以相应的规则保证状态以一种可预测的方式发生变化 通常 ...
- 1013A.Piles With Stones
题目出处:http://codeforces.com/contest/1013/problem/A #include<iostream> using namespace std; int ...
- AddressUtils
package com.ruoyi.common.utils; import org.slf4j.Logger; import org.slf4j.LoggerFactory; import com. ...
- 14 微服务电商【黑马乐优商城】:day04-项目搭建(一)
本项目的笔记和资料的Download,请点击这一句话自行获取. day01-springboot(理论篇) :day01-springboot(实践篇) day02-springcloud(理论篇一) ...
- regex(python)
正则表达式 #!/usr/bin/env python # -*- coding: utf-8 -*- # @Time : 2018/7/26 16:39 # @Author : jackendoff ...
- 在Myeclipse10中配置tomcat后新建工程
1.配置tomcat6.0 这里不在细说,和eclipse配置是一模一样的. 2.新建动态网站项目 3.配置显示服务器窗口 4.把项目与服务器链接 5.运行项目
- K-th K
题目描述 You are given an integer sequence x of length N. Determine if there exists an integer sequence ...
