bcftools
beftools非常复杂,大概有20个命令,每个命令下面还有N多个参数
annotate .. edit VCF files, add or remove annotations call .. SNP/indel calling (former "view") cnv .. Copy Number Variation caller concat .. concatenate VCF/BCF files from the same set of samples consensus .. create consensus sequence by applying VCF variants convert .. convert VCF/BCF to other formats and back csq .. haplotype aware consequence caller filter .. filter VCF/BCF files using fixed thresholds gtcheck .. check sample concordance, detect sample swaps and contamination index .. index VCF/BCF isec .. intersections of VCF/BCF files merge .. merge VCF/BCF files files from non-overlapping sample sets mpileup .. multi-way pileup producing genotype likelihoods norm .. normalize indels plugin .. run user-defined plugin polysomy .. detect contaminations and whole-chromosome aberrations query .. transform VCF/BCF into user-defined formats reheader .. modify VCF/BCF header, change sample names roh .. identify runs of homo/auto-zygosity sort .. sort VCF/BCF files stats .. produce VCF/BCF stats (former vcfcheck) view .. subset, filter and convert VCF and BCF files
下面讲一下过滤参数
1、bcftools filter(以-i参数为例)
- -i, --include EXPRESSION :
- include only sites for which EXPRESSION is true. For valid expressions see EXPRESSIONS.(根据正则保留)
- 其中包括:
- 1.1、numerical constants, string constants, file names (this is currently supported only to filter by the ID column)
1, 1.0, 1e-4 "String" @file_name
1.2、算术运算
+,*,-,/
1.3、comparison operators
== (same as =), >, >=, <=, <, !=
1.4、regex操作符“~”和它的否定“!~”。表达式区分大小写,除非添加“/i”。
INFO/HAYSTACK ~ "needle" INFO/HAYSTACK ~ "NEEDless/i"
1.5、圆括号
(, )
1.6、逻辑运算符。参见下面的示例和有关“&&”与“&”以及“||”与“|”之间区别的过滤教程。
&&, &, ||, |
1.7、信息标签,格式标签,列名
INFO/DP or DP FORMAT/DV, FMT/DV, or DV FILTER, QUAL, ID, CHROM, POS, REF, ALT[0]
1.8、1 (or 0) to test the presence (or absence) of a flag
FlagA=1 && FlagB=0
1.9、"." to test missing values
DP=".", DP!=".", ALT="."
2.0、missing genotypes can be matched regardless of phase and ploidy (".|.", "./.", ".") using these expressions
GT~"\.", GT!~"\."
2.1、sample genotype: reference (haploid or diploid), alternate (hom or het, haploid or diploid), missing genotype, homozygous, heterozygous, haploid, ref-ref hom, alt-alt hom, ref-alt het, alt-alt het, haploid ref, haploid alt (case-insensitive)
GT="ref" GT="alt" GT="mis" GT="hom" GT="het" GT="hap" GT="RR" GT="AA" GT="RA" or GT="AR" GT="Aa" or GT="aA" GT="R" GT="A"
2.2、TYPE for variant type in REF,ALT columns (indel,snp,mnp,ref,bnd,other,overlap). Use the regex operator "\~" to require at least one allele of the given type or the equal sign "=" to require that all alleles are of the given type. Compare
TYPE="snp" TYPE~"snp" TYPE!="snp" TYPE!~"snp"
2.3、array subscripts (0-based), "*" for any element, "-" to indicate a range. Note that for querying FORMAT vectors, the colon ":" can be used to select a sample and an element of the vector, as shown in the examples below
INFO/AF[0] > 0.3 .. first AF value bigger than 0.3 FORMAT/AD[0:0] > 30 .. first AD value of the first sample bigger than 30 FORMAT/AD[0:1] .. first sample, second AD value FORMAT/AD[1:0] .. second sample, first AD value DP4[*] == 0 .. any DP4 value FORMAT/DP[0] > 30 .. DP of the first sample bigger than 30 FORMAT/DP[1-3] > 10 .. samples 2-4 FORMAT/DP[1-] < 7 .. all samples but the first FORMAT/DP[0,2-4] > 20 .. samples 1, 3-5 FORMAT/AD[0:1] .. first sample, second AD field FORMAT/AD[0:*], AD[0:] or AD[0] .. first sample, any AD field FORMAT/AD[*:1] or AD[:1] .. any sample, second AD field (DP4[0]+DP4[1])/(DP4[2]+DP4[3]) > 0.3 CSQ[*] ~ "missense_variant.*deleterious"
2.4、with many samples it can be more practical to provide a file with sample names, one sample name per line
GT[@samples.txt]="het" & binom(AD)<0.01
2.5、function on FORMAT tags (over samples) and INFO tags (over vector fields): maximum; minimum; arithmetic mean (AVG is synonymous with MEAN); median; standard deviation; sum; string length; absolute value; number of elements (matching columns for FORMAT tags or number of fields for INFO tags).
MAX, MIN, AVG, MEAN, MEDIAN, STDEV, SUM, STRLEN, ABS, COUNT
2.6、two-tailed binomial test. Note that for N=0 the test evaluates to a missing value and when FORMAT/GT is used to determine the vector indices, it evaluates to 1 for homozygous genotypes.
binom(FMT/AD) .. GT can be used to determine the correct index binom(AD[0],AD[1]) .. or the fields can be given explicitly phred(binom()) .. the same as binom but phred-scaled
2.7、variables calculated on the fly if not present: number of alternate alleles; number of samples; count of alternate alleles; minor allele count (similar to AC but is always smaller than 0.5); frequency of alternate alleles (AF=AC/AN); frequency of minor alleles (MAF=MAC/AN); number of alleles in called genotypes; number of samples with missing genotype; fraction of samples with missing genotype; indel length (deletions negative, insertions positive)
N_ALT, N_SAMPLES, AC, MAC, AF, MAF, AN, N_MISSING, F_MISSING, ILEN
2.8、the number (N_PASS) or fraction (F_PASS) of samples which pass the expression
N_PASS(GQ>90 & GT!="mis") > 90 F_PASS(GQ>90 & GT!="mis") > 0.9
2.9、custom perl filtering. Note that this command is not compiled in by default, see the section Optional Compilation with Perl in the INSTALL file for help and misc/demo-flt.pl for a working example. The demo defined the perl subroutine "severity" which can be invoked from the command line as follows:
perl:path/to/script.pl; perl.severity(INFO/CSQ) > 3
注意事项:
字符串比较和正则表达式不区分大小写;变量和函数名不区分大小写,但是flag区分大小写。例如,"qual"可以代替"qual", "strlen()"可以代替"strlen()",但不是“dp”而是“DP”。当查询多个值时,将测试所有元素并对结果使用OR逻辑。例如,查询“TAG=1,2,3,4”时,计算如下:
-i 'TAG[*]=1' .. true, the record will be printed -i 'TAG[*]!=1' .. true -e 'TAG[*]=1' .. false, the record will be discarded -e 'TAG[*]!=1' .. false -i 'TAG[0]=1' .. true -i 'TAG[0]!=1' .. false -e 'TAG[0]=1' .. false -e 'TAG[0]!=1' .. true
举例:
MIN(DV)>5 MIN(DV/DP)>0.3 MIN(DP)>10 & MIN(DV)>3 FMT/DP>10 & FMT/GQ>10 .. both conditions must be satisfied within one sample FMT/DP>10 && FMT/GQ>10 .. the conditions can be satisfied in different samples QUAL>10 | FMT/GQ>10 .. true for sites with QUAL>10 or a sample with GQ>10, but selects only samples with GQ>10 QUAL>10 || FMT/GQ>10 .. true for sites with QUAL>10 or a sample with GQ>10, plus selects all samples at such sites TYPE="snp" && QUAL>=10 && (DP4[2]+DP4[3] > 2) COUNT(GT="hom")=0 .. no homozygous genotypes at the site AVG(GQ)>50 .. average (arithmetic mean) of genotype qualities bigger than 50 ID=@file .. selects lines with ID present in the file ID!=@~/file .. skip lines with ID present in the ~/file MAF[0]<0.05 .. select rare variants at 5% cutoff POS>=100 .. restrict your range query, e.g. 20:100-200 to strictly sites with POS in that range.
shell 扩展:
注意表达式必须经常引用,因为在shell中有些字符有特殊的含义。一个用单引号括起来的表达式的例子,它导致整个表达式按照预期传递给程序:
bcftools view -i '%ID!="." & MAF[0]<0.01'
------------------------过滤Filtering-------------
1、按照固定列(fixed columns)过滤
固定列,例如“QUAL, FILTER, INFO”可以直接过滤。例如:
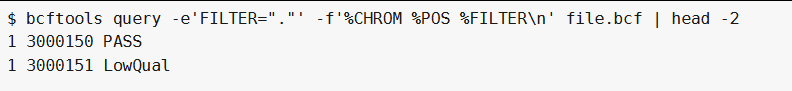
bcftools query -e'FILTER="."' -f'%CHROM %POS %FILTER\n' file.bcf #过滤掉FILTER字段中为.的行
bcftools query -i'QUAL>20 && DP>10' -f'%CHROM %POS %QUAL %DP\n' file.bcf | head -2 #只保留质量值大于20,且覆盖深度高于10的位点
2、FORMAT columns
在过滤FORMAT字段的时候,OR 逻辑用于所有samples。When filtering FORMAT tags, the OR logic is applied with multiple samples,而不是单个sample.例如,如果我们想删除任何样本中带有未知基因型的位点,表达式-i 'GT!="会不起作用,必须用相反的逻辑 -e 'GT ="."' :
bcftools query -i 'GT!="."' #不行 bcftools query -e 'GT ="."' #相反逻辑才可行

3、FORMAT列与 布尔值(&& vs & and || vs |)
我们希望一个sample或多个samples具有足够大的覆盖率(DP>10)和基因型质量(GQ>20)的snp位点:
bcftools query -i'FMT/DP>10 & FMT/GQ>20' -f'%POS[\t%SAMPLE:DP=%DP GQ=%GQ]\n' file.bcf ##-i 'FMT/DP>10和FMT/GQ>20'在同一个sample中选择满足条件的位点:

另一方面,如果我们需要在同一sample中两个条件都满足但不一定相同样品,我们使用&&操作符而不是&:
bcftools query -i'FMT/DP>10 && FMT/GQ>20' -f'%POS[\t%SAMPLE:DP=%DP GQ=%GQ]\n' file.bcf
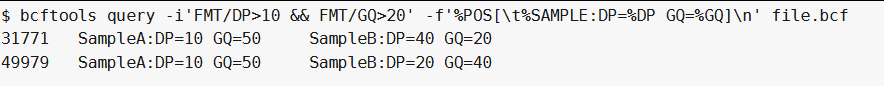
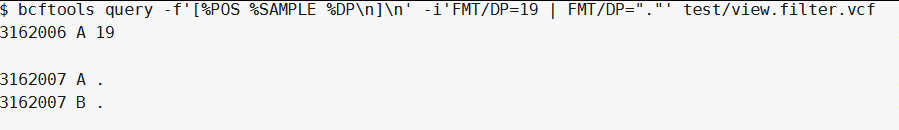
|操作符可以只选择匹配的样本:
bcftools query -f'[%POS %SAMPLE %DP\n]\n' -i 'FMT/DP=19 | FMT/DP="."' test/view.filter.vcf
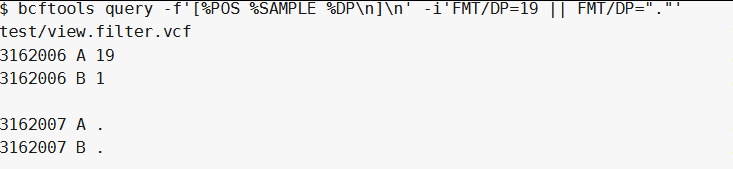
whole samples record when || is used(就是有一个样本符合该位点,那么该位点所有的样本记录都会被显示出来):
bcftools query -f '[%POS %SAMPLE %DP\n]\n' -i 'FMT/DP=19 || FMT/DP="."' test/view.filter.vcf
过滤:
bcftools filter -i 'SVLEN<100000 | SVLEN< -50 & DV>10' -Oz --threads 8 -o B1952.filter.clean.vcf.gz B1952.ngmlr_sniffle.vcf #例如过滤-50<绝对SVLEN<10000
重要网址:
http://samtools.github.io/bcftools/howtos/filtering.html
bcftools的更多相关文章
- bcftools合并vcf文件
见命令: bcftools merge A.vcf.gz B.vcf.gz C.vcf.gz -Oz -o ABC.vcf.gz 参考链接:http://vcftools.sourceforge.ne ...
- bcftools或vcftools提取指定区段的vcf文件(extract specified position )
下载安装bcftools 见如下命令: bcftools filter 1000Genomes.vcf.gz --regions 9:4700000-4800000 > 4700000-4800 ...
- 使用bcftools提取指定样本的vcf文件(extract specified samples in vcf format)
1.下载安装bcftools. 2.准备样本ID文件,这里命名为samplelistname.txt,一个样本一行,如下所示: sample1 sample2 sample3 3.输入命令: bcft ...
- bcftools将vcf生成bgzip和index格式
利用bcftools软件将vcf格式生成gz格式和index格式,需要用到“-Oz”和“index”命令,具体如下: /bcftools-1.8/bin/bcftools view ExAC.vcf ...
- 【Bcftools】合并不同sample的vcf文件,通过bcftools
通过GATK calling出来的SNP如果使用UnifiedGenotype获得的SNP文件是分sample的,但是如果使用vcftools或者ANGSD则需要Vcf文件是multi-sample的 ...
- 【BCFTOOLS】按样本拆分VCF文件
在对vcf的操作有这样三个软件: Vcftools:主要用于群体分析,文本处理的功能不是很强大,虽然这个软件也可以拆分样本,但是这种拆分不涉及文件的处理,只是保留在分析流程里. GATK .x:这个软 ...
- samtools+bcftools 进行SNP calling
两个软件的作用:1.samtools mpileup 主要是用于收集BAM文件中的信息,这个位点上有多少条read匹配,匹配read的碱基是什么,并将这些信息存储在BCF文件中.2.bcftools ...
- bcftools 提取vcf(snp/indel)文件子集
做群体变异检测后,通常会有提取子集的操作,之前没有发现bcftools有这个功能,都是自己写脚本操作,数据量一上来,速度真的是让人无语凝噎.这里记录下提取子vcf文件的用法,软件版本:bcftools ...
- linux 安装SAMtools,bcftools,htslib,sratoolkit,bedtools,GATK,TrimGalore,qualimap,vcftools,bwa
--------------------安装Samtools---------------------------------------------------------------------- ...
随机推荐
- Spring-AOP-配置实现五大通知
码云: xml配置方法:https://gitee.com/MarkPolaris/spring_aop_1 注解配置方法:https://gitee.com/MarkPolaris/spring-e ...
- 《Effective-Ruby》读书笔记
本篇是在我接触了 Ruby 很短一段时间后有幸捧起的一本书,下面结合自己的一些思考,来输出一下自己的读书笔记 前言 学习一门新的编程语言通常需要经过两个阶段: 第一个阶段是学习这门编程语言的语法和结构 ...
- Mybatis框架模糊查询+多条件查询
一.ISmbmsUserDao层 //根据姓名模糊查询 public List<Smbms> getUser(); //多条件查询 public List<Smbms> get ...
- Unity TextMeshPro替代Text组件创建简体中文字体纹理集
Unity原生的Text组件有一个毛病,只要文本放大字体放大就会有毛边或锯齿,一个更好的解决方案是用TextMeshPro替代ugui中的Text组件. TMPro采用SDF文字渲染技术,可以使文字放 ...
- 使用paramiko模块进行封装,远程操作linux主机
import time import paramiko class HandleParamiko: ''' 定义一个linux处理类 ''' def __init__(self, hostname, ...
- @PostConstruct - 静态方法调用IOC容器Bean对象
需求:工具类里面引用IOC容器Bean,强迫症患者在调用工具类时喜欢用静态方法的方式而非注入的方式去调用,但是spring 不支持注解注入静态成员变量. 静态变量/类变量不是对象的属性,而是一个类的属 ...
- Java入门系列之字符串创建方式、判断相等(一)
前言 陆续从0开始学习Java出于多掌握一门语言以后的路也会更宽,.NET和Java兼顾,虽然路还很艰难,但事在人为.由于Java和C#语法相似,所以关于一些很基础的内容不会再重头讲,Java系列中所 ...
- Python巧用法
#for 与 else 搭配使用(使用break跳过else) a=[1,2,3,4,5] for i in a: print(i) else: print(i, 'I am else!') for ...
- Xshell的一些使用方法和注意事项
xshell 本文就是想记录下最近遇到的一些问题,以及一些 xshell 能帮助我们提升效率的方面. xshell 编码问题 我们连接服务器,是通过本地登录到 跳板机,然后通过跳板机登录到 我们的服务 ...
- windows环境下安装配置MongoDB
版本选择MongoDB的版本命名规范如:x.y.z: y为奇数时表示当前版本为开发版,如:2.3.0.2.1.1: y为偶数时表示当前版本为稳定版,如:2.0.1.2.2.0: 目前官网上最新的版本为 ...

