吴裕雄--天生自然 PYTHON数据分析:医疗数据分析
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv) # plotly
import chart_studio.plotly as py
from plotly.offline import init_notebook_mode, iplot
init_notebook_mode(connected=True)
import plotly.graph_objs as go
import seaborn as sns
# word cloud library
from wordcloud import WordCloud # matplotlib
import matplotlib.pyplot as plt
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
dataframe = pd.read_csv("F:\\kaggleDataSet\\healthcare-data\\test_2v.csv")
import chart_studio.plotly as py
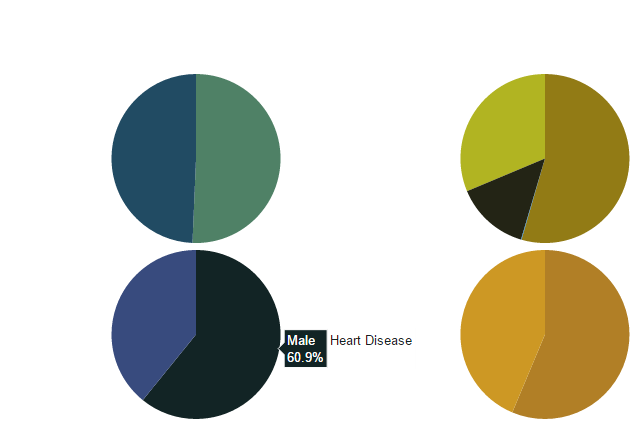
from plotly.graph_objs import * df_heart_disease = dataframe[dataframe.heart_disease== 1]
labels = df_heart_disease.gender
pie1_list=df_heart_disease.heart_disease df_hypertension= dataframe[dataframe.hypertension == 1]
labels1 = df_hypertension.gender
pie1_list1=df_hypertension.hypertension labels2 = dataframe.Residence_type
pie1_list2 = dataframe.heart_disease labels3 = dataframe.work_type
pie1_list3 = dataframe.heart_disease fig = {
'data': [
{
'labels': labels,
'values': pie1_list,
'type': 'pie',
'name': 'Heart Disease',
'marker': {'colors': ['rgb(56, 75, 126)',
'rgb(18, 36, 37)',
'rgb(34, 53, 101)',
'rgb(36, 55, 57)',
'rgb(6, 4, 4)']},
'domain': {'x': [0, .48],
'y': [0, .49]},
'hoverinfo':'label+percent+name',
'textinfo':'none'
},
{
'labels': labels1,
'values': pie1_list1,
'marker': {'colors': ['rgb(177, 127, 38)',
'rgb(205, 152, 36)',
'rgb(99, 79, 37)',
'rgb(129, 180, 179)',
'rgb(124, 103, 37)']},
'type': 'pie',
'name': 'Hypertension',
'domain': {'x': [.52, 1],
'y': [0, .49]},
'hoverinfo':'label+percent+name',
'textinfo':'none' },
{
'labels': labels2,
'values': pie1_list2,
'marker': {'colors': ['rgb(33, 75, 99)',
'rgb(79, 129, 102)',
'rgb(151, 179, 100)',
'rgb(175, 49, 35)',
'rgb(36, 73, 147)']},
'type': 'pie',
'name': 'Residence Type',
'domain': {'x': [0, .48],
'y': [.51, 1]},
'hoverinfo':'label+percent+name',
'textinfo':'none'
},
{
'labels': labels3,
'values': pie1_list3,
'marker': {'colors': ['rgb(146, 123, 21)',
'rgb(177, 180, 34)',
'rgb(206, 206, 40)',
'rgb(175, 51, 21)',
'rgb(35, 36, 21)']},
'type': 'pie',
'name':'Work Type',
'domain': {'x': [.52, 1],
'y': [.51, 1]},
'hoverinfo':'label+percent+name',
'textinfo':'none'
} ],
'layout': {'title': '',
'showlegend': False}
} iplot(fig)
import chart_studio.plotly as py
import plotly.graph_objs as go # Create random data with numpy
import numpy as np df_250 = dataframe.iloc[:250,:] random_x = df_250.index
random_y0 = df_250.avg_glucose_level
random_y1 = df_250.bmi
random_y2 = df_250.age # Create traces
trace0 = go.Scatter(
x = random_x,
y = random_y0,
mode = 'markers',
name = 'Avg. Glucose Level'
)
trace1 = go.Scatter(
x = random_x,
y = random_y1,
mode = 'lines+markers',
name = 'BMI'
)
trace2 = go.Scatter(
x = random_x,
y = random_y2,
mode = 'lines',
name = 'Age'
) data = [trace0, trace1, trace2]
iplot(data, filename='scatter-mode')

import chart_studio.plotly as py
import plotly.graph_objs as go
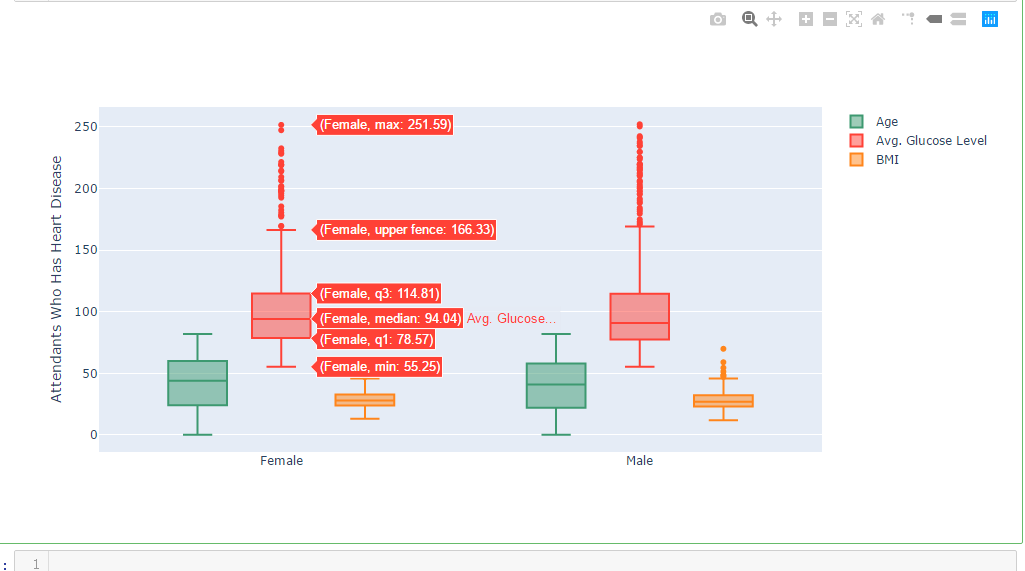
df_heart_disease = dataframe[dataframe.heart_disease==1]
labels = df_heart_disease.gender
x = labels trace0 = go.Box(
y=dataframe.age,
x=x,
name='Age',
marker=dict(
color='#3D9970'
)
)
trace1 = go.Box(
y=dataframe.avg_glucose_level,
x=x,
name='Avg. Glucose Level',
marker=dict(
color='#FF4136'
)
)
trace2 = go.Box(
y=dataframe.bmi,
x=x,
name='BMI',
marker=dict(
color='#FF851B'
)
)
data = [trace0, trace1, trace2]
layout = go.Layout(
yaxis=dict(
title='Attendants Who Has Heart Disease',
zeroline=False
),
boxmode='group'
)
fig = go.Figure(data=data, layout=layout)
iplot(fig)
import chart_studio.plotly as py
import plotly.graph_objs as go
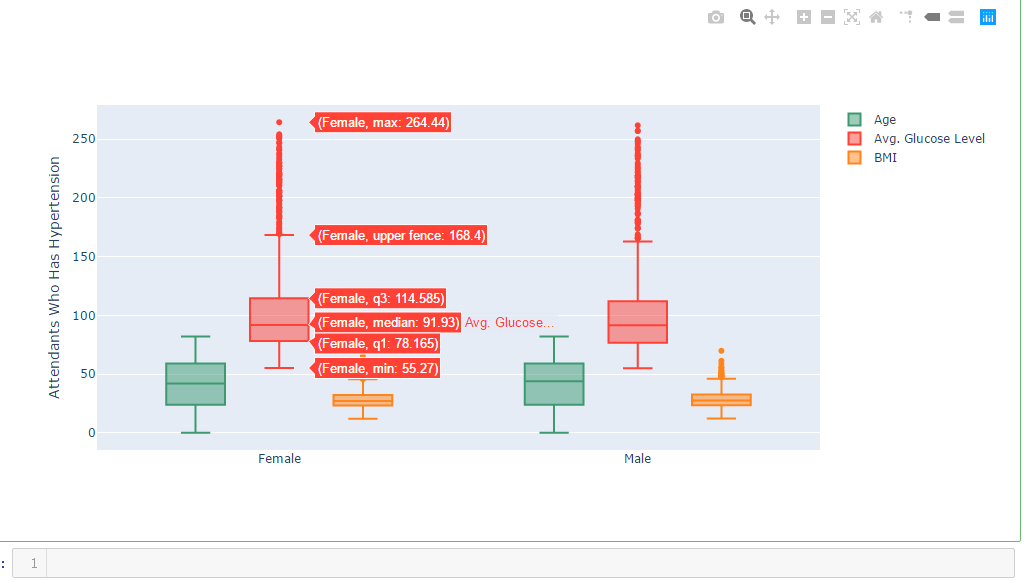
df_hypertension= dataframe[dataframe.hypertension == 1]
labels1 = df_hypertension.gender
x = labels1 trace0 = go.Box(
y=dataframe.age,
x=x,
name='Age',
marker=dict(
color='#3D9970'
)
)
trace1 = go.Box(
y=dataframe.avg_glucose_level,
x=x,
name='Avg. Glucose Level',
marker=dict(
color='#FF4136'
)
)
trace2 = go.Box(
y=dataframe.bmi,
x=x,
name='BMI',
marker=dict(
color='#FF851B'
)
)
data = [trace0, trace1, trace2]
layout = go.Layout(
yaxis=dict(
title='Attendants Who Has Hypertension',
zeroline=False
),
boxmode='group'
)
fig = go.Figure(data=data, layout=layout)
iplot(fig)
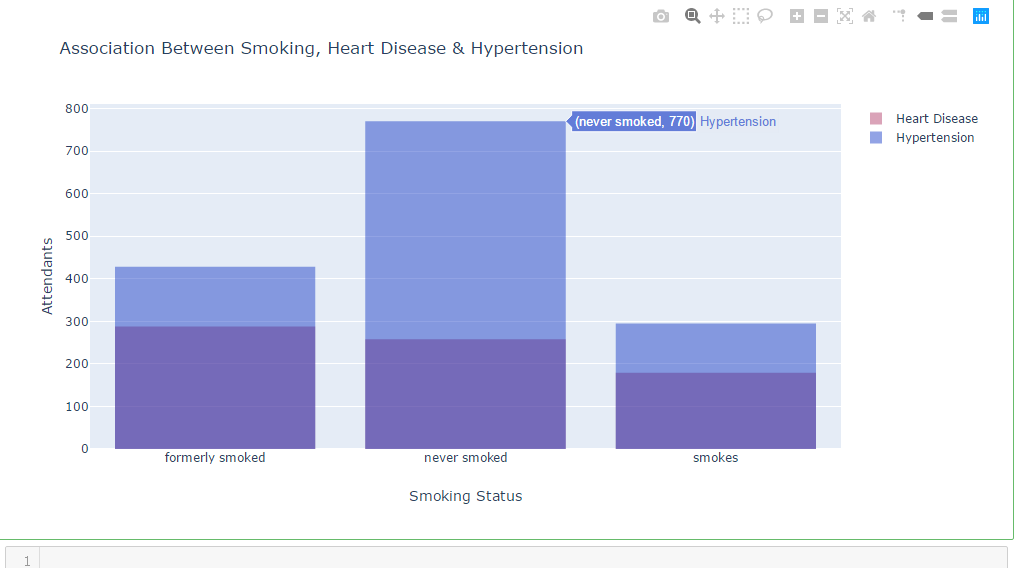
df_heart_disease_1 = dataframe.smoking_status [dataframe.heart_disease == 1 ]
df_hypertension_1 = dataframe.smoking_status [dataframe.hypertension == 1 ]
trace1 = go.Histogram(
x=df_heart_disease_1,
opacity=0.75,
name = "Heart Disease",
marker=dict(color='rgba(171, 50, 96, 0.6)'))
trace2 = go.Histogram(
x=df_hypertension_1,
opacity=0.75,
name = "Hypertension",
marker=dict(color='rgba(12, 50, 196, 0.6)')) data = [trace1, trace2]
layout = go.Layout(barmode='overlay',
title=' Association Between Smoking, Heart Disease & Hypertension',
xaxis=dict(title='Smoking Status'),
yaxis=dict( title='Attendants'),
)
fig = go.Figure(data=data, layout=layout)
iplot(fig)
df_heart_disease_1 = dataframe.work_type [dataframe.heart_disease == 1 ]
df_hypertension_1 = dataframe.work_type [dataframe.hypertension == 1 ] trace1 = go.Histogram(
x=df_heart_disease_1,
opacity=0.75,
name = "Heart Disease",
marker=dict(color='rgba(171, 50, 96, 0.6)'))
trace2 = go.Histogram(
x=df_hypertension_1,
opacity=0.75,
name = "Hypertension",
marker=dict(color='rgba(12, 50, 196, 0.6)')) data = [trace1, trace2]
layout = go.Layout(barmode='overlay',
title=' Association Between Work Type, Heart Disease & Hypertension',
xaxis=dict(title=''),
yaxis=dict( title='Attendants'),
)
fig = go.Figure(data=data, layout=layout)
iplot(fig)

吴裕雄--天生自然 PYTHON数据分析:医疗数据分析的更多相关文章
- 吴裕雄--天生自然 PYTHON数据分析:糖尿病视网膜病变数据分析(完整版)
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 PYTHON数据分析:所有美国股票和etf的历史日价格和成交量分析
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 python数据分析:健康指标聚集分析(健康分析)
# This Python 3 environment comes with many helpful analytics libraries installed # It is defined by ...
- 吴裕雄--天生自然 python数据分析:葡萄酒分析
# import pandas import pandas as pd # creating a DataFrame pd.DataFrame({'Yes': [50, 31], 'No': [101 ...
- 吴裕雄--天生自然 PYTHON数据分析:人类发展报告——HDI, GDI,健康,全球人口数据数据分析
import pandas as pd # Data analysis import numpy as np #Data analysis import seaborn as sns # Data v ...
- 吴裕雄--天生自然 python数据分析:医疗费数据分析
import numpy as np import pandas as pd import os import matplotlib.pyplot as pl import seaborn as sn ...
- 吴裕雄--天生自然 PYTHON语言数据分析:ESA的火星快车操作数据集分析
import os import numpy as np import pandas as pd from datetime import datetime import matplotlib imp ...
- 吴裕雄--天生自然 python语言数据分析:开普勒系外行星搜索结果分析
import pandas as pd pd.DataFrame({'Yes': [50, 21], 'No': [131, 2]}) pd.DataFrame({'Bob': ['I liked i ...
- 吴裕雄--天生自然 PYTHON数据分析:基于Keras的CNN分析太空深处寻找系外行星数据
#We import libraries for linear algebra, graphs, and evaluation of results import numpy as np import ...
随机推荐
- SRS|Stratified sampling|系统抽样|Cluster sampling|multistage sampling|
生物统计学 总体和抽样 抽样方法: ========================================================= 简单随机抽样SRS:随机误差,系统误差 标准误, ...
- ansible puppet saltstack三款自动化运维工具的对比
一.基础介绍 ansible基础介绍可参考:http://www.linuxidc.com/Linux/2017-12/149671.htm puppet基础介绍可参考:http://www.linu ...
- 4.docker 简介
1.概念 docker 提供了一个开发 打包 运行 app 的平台 通过 docker engine 把 app 和底层infrastructure隔离开来 2.docker engine 所包含的内 ...
- StartDT AI Lab | 智能运筹助力企业提升决策效率、优化决策质量
在人工智能和大数据时代,越来越多的云上数据和越来越智能的模型开始辅助人们做出各种最优决策,从运营效率.成本节约.最优配置等方方面面,实现降本增效,进一步提升商业效率.京东.美团.滴滴.顺丰等众多知名厂 ...
- D - Daydreaming Stockbroker Gym - 101550D
题目链接:http://codeforces.com/gym/101550/attachments 总的来说就是要: 极大值卖出,极小值买入, 再加上端点时的特判. 还有就是会有连续几天股票价格相同的 ...
- Java和Mysql中的数据类型
1.mysql中的基本类型 1.整数: tinyint:1个字节 -128~127 smallint: 2个字节 -32768~32767 int : 4个字节 bigint: 8个字节 2 ...
- 尝试brpc来升级rpc服务,测试应用过程
照着官方文档来,不过在mac下还是有些小坑 对熟悉c++的人来说很小儿科,但对c++相对比较外行 (只知道基本语法和部分数据结构)的人,还是作不到开箱即用 首先编译```If you need to ...
- Opencv笔记(十二)——形态学转换
学习目标: 学习不同的形态学操作,例如腐蚀,膨胀,开运算,闭运算等 我们要学习的函数有: cv2.erode(), cv2.dilate(), cv2.morphologyEx()等 原理简介: 形态 ...
- 项目中关于RPC 和rocketMQ使用场景的感受
在花生待的这半年,切身体会了系统之间交互场景的接口技术实现方式,个人总结.仅供参考: 1.关于rpc接口,一般情况下 都是同步的.A系统的流程调用B系统.等着B返回,根据返回结果继续进行A接下来的流程 ...
- CodeForces 527C. Glass Carving (SBT,线段树,set,最长连续0)
原题地址:http://codeforces.com/problemset/problem/527/C Examples input H V V V output input H V V H V ou ...
